Load FindZebra Summary
Disclaimer:
FindZebra Search conducts a search using our specialized medical search engine.
FindZebra Summary uses the text completions API
(subject to OpenAI’s API data usage policies)
to summarize and reason about the search results.
The search is conducted in publicly available information on the Internet that we present “as is”.
You should be aware that FindZebra is not supplying any of the content in the search results.
FindZebra Summary is loading...
-
Pancreatic Neuroendocrine Tumor
GARD
However in some cases, a pancreatic NET occurs outside of the pancreas. A NET arises from cells that produce hormones, so the tumor can also produce hormones. ... Pancreatic NETs are called either functional or nonfunctional. A functional pancreatic NET causes specific symptoms because it makes extra hormones, such as gastrin, insulin, or glucagon. ... Pancreatic NETs can be hard to diagnosis, often not identified until 5 to 10 years after they begin to grow. Most pancreatic NETs are not inherited and occur sporadically in people with no family history of NETs.MEN1, PCSK1, ATM, BRCA2, C11orf65, IGF2, SST, TP53, CDKN2A, SLC6A2, MTOR, EPHB1, POMC, GH1, GCGR, DAXX, ELK3, KRT19, SSTR2, CHGA, SSTR5, UCHL1, FZD4, GCM2, DLGAP1, DCLK1, SSTR4, INA, STK11, EIF2AK3, TFE3, THBD, CXCR4, PAX8, TSC1, TTR, TYMS, VEGFA, ABO, CNPY2, MRGPRX4, GPR166P, VN1R17P, MIR196A1, GADL1, MRGPRX1, GPRC6A, OXER1, GPR119, GPR151, MRGPRX3, SEMA3A, AZIN2, ACCS, STK33, LGR6, ACSS2, MEG3, NEUROG3, LPAR3, LILRB1, PLA2G15, RET, SLC2A3, INSM1, GRN, FFAR1, GHRH, GAST, FGFR4, F3, EGFR, DHCR24, CSF1, CRH, CHGB, CD44, CCK, CALCA, VPS51, ATRX, ASS1, ASCL1, ANGPT2, HSF1, PDX1, SLC2A2, KIT, SLC2A1, SEA, SDHB, SDHA, AKT1, PYGM, PTH, PTEN, PPY, PTPA, PGR, PCYT1A, PCNA, NFKB1, NEUROD1, MUC1, SMAD4, STMN1, KRAS, H3P10
-
Uric Acid Concentration, Serum, Quantitative Trait Locus 1
OMIM
The findings were replicated in the ARIC cohort of 11,024 white and 3,843 black individuals, yielding p values of 9.7 x 10(-30) and 9.8 x 10(-4), respectively. The combined p value for white individuals from all 3 cohorts was 2.5 x 10(-60), and further analysis showed that the SNP was direction-consistent with the development of gout in white participants (OR of 1.74; p = 3.3 x 10(-15)). ... Evidence for both an increased rate of uric acid synthesis and an impaired net elimination of uric acid by the kidney has been advanced. ... The Q126X allele was associated with a significantly increased risk of hyperuricemia (odds ratio (OR) of 3.61; p = 2.91 x 10(-7)) and gout (OR of 4.25, p = 3.04 x 10(-8)). The Q141K allele was associated with a significantly increased risk of hyperuricemia (OR of 2.06, p = 1.53 x 10(-11)) and gout (OR of 2.23; p = 5.54 x 10(-11)).
-
Chronic Hallucinatory Psychosis
Wikipedia
Chronic Hallucinatory Psychosis Specialty Psychiatry Chronic hallucinatory psychosis is a psychosis subtype, classified under "Other nonorganic psychosis" by the ICD-10 Chapter V: Mental and behavioural disorders . ... Others, again, might be swept into the widespread net of dementia praecox . This state of affairs cannot be regarded as satisfactory, for they are not truly cases of melancholia, paranoia, dementia praecox or any other described affection. ... Also, it needs to be noted that the delusion is a comparatively late arrival and is the logical result of the hallucinations. [1] References [ edit ] ^ A paper read at the Quarterly Meeting of the Medico-Psychological Association on February 24th, 1920, written by Robert Hunter Steen, King's College Hospital, London External links [ edit ] Classification D ICD - 10 : F28
-
Neuroendocrine Tumor
Wikipedia
H&E stain Specialty Endocrine oncology Neuroendocrine tumors ( NETs ) are neoplasms that arise from cells of the endocrine ( hormonal ) and nervous systems . ... G1 and G2 neuroendocrine neoplasms are called neuroendocrine tumors (NETs) – formerly called carcinoid tumours. ... Unsourced material may be challenged and removed. ( November 2015 ) ( Learn how and when to remove this template message ) NETs from a particular anatomical origin often show similar behavior as a group, such as the foregut (which conceptually includes pancreas, and even thymus, airway and lung NETs), midgut and hindgut ; individual tumors within these sites can differ from these group benchmarks: Foregut NETs are argentaffin negative. ... Bone metastasis is uncommon. Hindgut NETs are argentaffin negative and rarely secrete 5-HT, 5-HTP, or any other vasoactive peptides. ... Not all cells are immediately killed; cell death can go on for up to two years. [ citation needed ] PRRT was initially used for low grade NETs. It is also very useful in more aggressive NETs such as Grade 2 and 3 NETs [83] [84] provided they demonstrate high uptake on SSTR imaging to suggest benefit.MEN1, CDKN1B, SSTR2, DAXX, ATRX, BRAF, TYMS, PTHLH, SSTR3, SSTR1, BAP1, MTOR, SST, GAST, SLC6A2, INSM1, CTNNB1, RET, PIK3CA, DNMT3A, POMC, EPHB1, PIK3CG, PIK3CD, CHGA, ELK3, CHEK2, PIK3CB, GRN, CD274, SMUG1, AKT1, GNA12, TP53, SYP, VEGFA, CDKN2A, ASCL1, BCL2, ENO2, NCAM1, GCG, MYCN, EGFR, MGMT, KIT, RASSF1, VHL, SCLC1, SSTR5, FOLH1, NKX2-1, KRAS, CALCA, CCND1, TAC1, PTPRF, VIP, NTS, PAX5, RHBDF2, GRP, IGF1, SDHD, GOT1, MAP2K7, CCK, ERBB2, DLL3, PPY, CXCL12, TP63, SMAD4, MUC1, INS, GCGR, CKAP4, NEUROD1, ISL1, MYC, NGF, SATB2, GLP1R, HSP90AA1, H3P10, HRAS, CHGB, CALR, NTRK1, TEK, DLK1, CDK4, CDX2, TGFA, UCHL1, RPE65, PGR, PDGFRA, CARTPT, CRH, UVRAG, SLC5A5, CXCR4, IGF1R, OTP, IL6, PHLDA3, TTF1, PAX8, TACR1, STK11, TRIM21, PLA2G15, SCG2, SQLE, SLC18A2, TERT, HDAC9, SLC2A1, PROM1, BCL2L11, NTSR1, PAX6, NAMPT, NOCT, INA, PLCB3, CD200, MKI67, PDX1, MAPK1, NES, HPSE, PTEN, STMN1, ABO, RIPK1, RORC, RAF1, IL1B, TRPV1, GATA3, ANGPT2, FOXM1, PTK2B, SDHAF2, ACCS, BDNF, EPAS1, EGF, ACSS2, MIB1, DNMT1, CCN2, TRPM8, CLDN4, CPE, CD34, CD44, FLNA, CEACAM5, B3GAT1, GH1, GIP, GHSR, GIPR, ADCY2, ALB, H3P28, TPPP2, H4C5, GGH, MIR1290, TMEM209, ELOA3, H4C13, H4C14, GPR151, SRPX, LGR5, TNFSF11, PSMG1, DCBLD2, H4-16, NRP1, MRGPRX4, SOCS1, H4C2, MIR3137, MRGPRX3, TNFRSF25, H3P12, CYYR1, AZIN2, DNER, AK6, MLIP, LMLN, NRP2, GPR68, MIR1246, H4C8, MAFK, MIR150, MIR155, MBOAT4, H4C9, MIR21, POTEKP, VN1R17P, SNORD95, GPR166P, ARID1A, EID3, SLC7A5, MIR375, H4C15, FZD4, MIRLET7C, OXER1, H4C12, HMGA2, H4C3, ARX, ELOA3B, GPRC6A, H4C11, H4C6, C17orf97, POTEM, MRGPRX1, ARMH1, H4C1, GADL1, ACTBL2, H4C4, BRI3, SQSTM1, ISYNA1, GHRL, ACOT7, KLF12, KRT20, SLC27A4, TET2, BCOR, EBNA1BP2, RALBP1, PGRMC1, LAMTOR1, FBXW7, MEG3, MAML3, TMEM127, NTNG1, ATRAID, KHDRBS1, DCTN4, SNORD61, NUP62, SNORD48, NTSR2, LPAR3, MAPK8IP2, SRRM2, BRD4, TRAM1, SPINK4, XIST, PPWD1, RBMS3, SETD1B, ZHX2, TNFSF13B, USE1, MAK16, UBE2Z, ONECUT2, FHL5, GCM2, DCLK1, ZBED1, ARHGEF2, PALB2, ALG9, SNED1, TET1, PDCD1LG2, TMPRSS13, MTA1, RPAIN, H1-10, EEF1E1, LGR6, PRMT5, NEUROD4, YAP1, SCML2, LANCL1, PAK4, RABEPK, ZNF197, CTNNBL1, PNO1, INSL5, EPB41L5, HDAC5, AKT3, CD302, GBA3, DCAF1, ATAT1, SERPINA3, VCL, CGA, ESR1, ERBB4, EPHB2, E2F1, DUSP2, DSG3, DPT, DPP4, DMBT1, DDC, DAD1, VCAN, CREB1, CRABP1, KLF6, CLU, FOXN3, CEACAM7, CEACAM3, ESR2, ETFA, EZH2, GHRH, HSPA4, AGFG1, HMOX1, HMGA1, GTF2H1, GSN, GNAS, GNA15, GFRA1, F3, GDNF, FSHR, FLT4, FLII, FLI1, FOXO1, FHIT, FGFR4, CGB3, CFL1, UQCRFS1, CDKN2C, FAS, APRT, APLP1, XIAP, APC, SLC25A6, SLC25A4, ANGPT1, ALK, AKT2, AFP, PARP1, ADCYAP1R1, ADCYAP1, ACVRL1, ACTN4, ACTG2, ACTG1, ACR, AQP4, ARF1, ATM, CASP3, CDK6, CD40LG, CD36, CD33, CCNE1, CCKBR, SERPINA6, CAV1, CA9, ATOH1, VPS51, C5, BRS3, BRCA2, DST, BAX, AVP, ATP4A, HTC2, HTR2A, TNC, IAPP, SDC1, SCT, SORT1, RNASE3, RARB, PTPRZ1, PTPRM, PTBP1, PSMD7, PSG2, PRKAR1A, PPP4C, POU4F1, PNN, PKD2, PITX2, PCYT1A, SERPINA5, PAX4, SDCBP, SDHB, SDHC, ST2, UBE2I, TPM3, TPH1, TNF, TM7SF2, TERC, TAT, STAT3, SSTR4, SEMA3F, SSR2, SOX11, SOX4, SOX2, SLPI, SLC3A2, SLC1A5, SFRP1, PAK3, PAK1, TNFRSF11B, KIF11, MDK, MAOA, LCN2, RPSA, L1CAM, KRT19, KRT7, KRT5, IL12A, MET, IL9, CXCL8, IL2, IL1A, IGFBP1, IGF2, IFNA13, IFNA1, MDM2, MFAP1, ODC1, MUTYH, NTRK2, NT5E, NRAS, NOTCH3, NPY, NOTCH1, NFKB1, NEFM, MUC4, CD99, NUDT1, COX2, MTAP, MST1R, MST1, MSMB, MMP7, MLH1, PTPRC
-
Epilepsy, Progressive Myoclonic, 10
OMIM
A number sign (#) is used with this entry because of evidence that progressive myoclonic epilepsy-10 (EPM10) is caused by homozygous mutation in the PRDM8 gene (616639) on chromosome 4q21. ... Description Progressive myoclonic epilepsy-10 is an autosomal recessive neurodegenerative disorder characterized by onset of progressive myoclonus, ataxia, spasticity, dysarthria, and cognitive decline in the first decade of life. ... Studies in patient tissues showed no net change in glycogen synthase (see, e.g., GYS1, 138570) activity, indicating that polyglucosan formation was not related to glycogen synthase.
-
Pancreatic Cancer
Wikipedia
NETs can start in most organs of the body, including the pancreas, where the various malignant types are all considered to be rare . ... Most cases occur after age 65, [10] while cases before age 40 are uncommon. The disease is slightly more common in men than in women. [10] In the United States, it is over 1.5 times more common in African Americans , though incidence in Africa is low. [10] Cigarette smoking is the best-established avoidable risk factor for pancreatic cancer, approximately doubling risk among long-term smokers, the risk increasing with the number of cigarettes smoked and the years of smoking. ... These are macroscopic lesions, which are found in about 2% of all adults. This rate rises to about 10% by age 70. These lesions have about a 25% risk of developing into invasive cancer. ... Typical sites for metastatic spread (stage IV disease) are the liver, peritoneal cavity and lungs , all of which occur in 50% or more of fully advanced cases. [58] PanNETs [ edit ] The 2010 WHO classification of tumors of the digestive system grades all the pancreatic neuroendocrine tumors (PanNETs) into three categories, based on their degree of cellular differentiation (from "NET G1" through to the poorly differentiated "NET G3"). [19] The U.S.BRCA2, TP53, CDKN2A, STK11, KRAS, BRCA1, SMAD4, EP300, PALB2, ABO, NR5A2, MYC, CTNNB1, TNF, MMP2, AKT2, APC, PDX1, PTEN, PTGS2, STAT3, ATM, KDR, CXCL8, TYMS, SST, TNFSF10, TERT, EGFR, TGFB1, HIF1A, MSLN, CD44, SPINK1, MMP9, PPARG, CD24, LDHA, PLAU, SOD2, PALLD, SSTR2, SLC29A1, MAP2K4, DPYD, TGM2, AXL, IL24, IFNA1, PTCH1, EPCAM, NR4A1, TFPI2, CDH1, LINC00673, NRP1, RAP1GAP, MEN1, RB1, VHL, WT1, KLF5, TYMP, PTHLH, GADD45A, FGF13, CCL20, EFEMP1, CFLAR, AHR, TNFRSF10B, SSTR1, FXYD3, TP63, BAP1, FOXP1, ANXA10, DAXX, NFKBIA, TSC2, RNF43, BUB1, VEGFB, MIR1179, SSTR3, IFNA2, RABL3, SUGCT, CNR1, IFNA5, CNR2, ETAA1, CLPTM1, SYCP1, RBBP8, GAGE1, CCAT1, BCL2L1, BCAR1, LY6E, ZNRF3, ATRX, COL18A1, CLPTM1L, DAB2, SBF2, HSPB2, ERBB2, CCKBR, HRAS, HSPB1, HSP90AA1, LINC01829, PPFIBP1, EPHB2, PIK3CA, CCK, HGF, ERBB3, MIA, PDHX, PSCA, SIRT1, HSPB3, H3P10, PIM3, NOTCH1, CEACAM5, EGF, SMUG1, PIK3CB, PARP1, MAPK1, MAPK8, MTCO2P12, GPC1, CDK4, HDAC9, CEACAM3, ABCG2, ACTB, CEACAM7, LINC01088, MAPK3, NDRG1, PSG2, YAP1, PIK3CG, TAFA5, SLC19A1, SPARC, PRSS1, PROM1, PIK3CD, HP, GLI1, MAP2K7, F3, NGF, ZEB1, FBXW7, NFE2L2, TEX11, ITGB1, VEGFA, GABPA, BICD1, ANXA2, CD82, GAST, DCLK1, COX2, CCND1, XIAP, MET, BACH1, S100A4, LGALS1, MDM2, DPP6, MUC16, FOXM1, MIR21, SLC39A4, MIR221, IFNG, EZH2, LINC00964, AKT1, ALB, LDLRAD4, C9, MUC5AC, SETD2, CXCR4, VIM, MUC4, CD274, LEPQTL1, XRCC1, IGF1, MUC1, IGF1R, CXCL12, IL1B, IL6, BCL2, HNF1A, SCT, TWIST1, CCN2, TLR4, DNMT1, AURKA, CAV1, PLK1, CASP3, PKM, MTHFR, LGALS3, MTOR, BRAF, PRKAA1, MIR301A, AGER, HOTAIR, EPHA2, MCL1, VTCN1, COL11A2, RAC1, DUSP6, CDKN2B, CRP, MAPK14, RUNX3, MALAT1, FN1, MIR155, MIR210, ERCC2, DCK, AGR2, CFTR, GDF15, CDKN1A, ADIPOQ, PRKN, MIR200C, TIMP1, KIF20A, PTF1A, PSMD9, TGFBR2, TICAM2, RRM1, MIR137, MIR142, ALOX5, MLH1, MIR29A, SOCS3, IFI27, SRC, GLP1R, PRKAA2, SP1, SOX2, TMED7-TICAM2, HMGA1, SOAT1, H3P23, PAK4, CHEK1, ZNRD2, BMI1, HMGA2, RALBP1, PALD1, TMED7, TLR7, BSG, DCTN6, WNT5A, EPAS1, WEE1, POSTN, BNIP3, MST1, FGFR1, ATN1, MST1R, GSK3B, PRKAB1, CD47, NFATC2, HSPA5, FANCG, PVT1, SHH, MBD1, ETS1, NPTX2, ABCB1, SLC2A1, MUC2, IFNA13, MMP7, MRC1, IGF2, CCN1, NQO1, LUM, MDK, IL10, PTPA, ELAVL2, POU5F1, FLT1, CXCR2, GSTT1, AIMP2, DKK3, MIR23A, KLF10, CRK, MIR214, MUL1, MIR203A, MIR200B, MIR195, APEX1, GRAP2, MIR145, CDKN1B, MIR141, RIPK1, TNFRSF6B, NR1I2, LINC01194, TGFA, THBS1, ALDH1A1, POLDIP2, MICA, NES, HOTTIP, UCA1, POU5F1P4, MIR34A, POU5F1P3, RNF19A, CASP8, CBLL2, CTLA4, AHSA1, RUNX2, ERCC1, OGG1, FANCC, ZG16B, DNER, NTRK1, FGF2, F2RL1, MMP14, FOXO3, CD276, NUPR1, RMC1, MSH2, AFAP1-AS1, DCC, LIF, MIR10B, HDAC1, HDAC2, HMGB1, ACTN4, HMOX1, ADM, ADRB2, IAPP, ICAM1, GDNF, MIR96, MIR25, IL1A, IL2, MIR223, MIR212, MIR205, ANXA1, MIR148A, MMRN1, MIR143, LCN2, MIR132, REG3A, TMEM97, UQCRFS1, STAT1, RET, TGFBI, CDC42, DPP4, CDK2, KLF4, CTNND1, IL32, DSPP, BAG3, CUX1, VDR, TAZ, PIM1, PCNA, SOX9, SEMA5A, XPO1, MAP2K1, SUB1, PRKD1, SPP1, JUN, TOP2A, MLRL, XIST, RAF1, IL4, IL4R, VEGFC, MIR222, MIR216A, NAT2, IL13, MOK, RAD51, TP73, JAK2, MIR23B, MIR29C, IGFBP3, MIR506, SMARCA1, SLC5A5, H3P9, COMMD3-BMI1, SNAI1, HK2, HLA-A, SKP2, SSTR5, FOXA2, SDC1, HSF1, CCL5, MIR27A, MIR494, MIR429, TNC, RRM2, MIR335, MIR17HG, TRA, RREB1, ROCK1, MIR30A, L1CAM, TEAD1, NCOA3, IKZF3, STMN1, BHLHE40, ESRP1, NT5C3A, IL22, PLEC, NXT1, SLC22A3, IL17B, ZEB2, NF2, NFATC1, TSPAN1, CIB1, CACYBP, MRPL28, PDPN, NFKB1, WWTR1, IGF2BP3, BRD4, HPSE, NRAS, NTSR1, COPS5, SERPINE1, PAK1, SIRT3, CDK14, PARPBP, PACC1, MSX2, TP53INP1, SOCS1, LEP, TBC1D9, MIR107, LGALS4, DUSP28, MTA1, PRKCA, UPRT, TM4SF1, LPAR2, SMAD2, MAGT1, MUC13, REG4, VMP1, LONP1, EHMT1, MMP1, FTO, TRPM8, GAS5, NDRG2, KEAP1, ACKR3, SLC12A9, ADAM9, NAT1, CDK6, GEM, GCG, CALR, CDKN3, FGFR2, ADH1C, CCR6, GLS, FHIT, CSF2, FOS, ALDH2, ACVR1B, FKBP5, FOXO1, ALPP, ALPI, GATA6, CETN1, ASS1, TIMM8A, CLDN7, F2R, TSACC, GPRC5A, ADH1B, HULC, FOSB, CASR, PCLAF, SMARCA4, DECR1, CDX2, CDK5, EPHX2, RCBTB1, FSCN1, SOX4, FLT3, MIR663A, LGALS9, UGT1A7, MIR217, CEL, CYP17A1, CLDN1, TRPM7, ISG15, CLOCK, MIR345, SET, CDK7, MIR200A, MIR216B, JUNB, JUND, ADAMTS1, KL, ITGAM, SERPINF1, SFTPD, SHC1, SELE, ITGA2, HDAC3, ARNTL, PBK, TUG1, SLC16A1, PDGFRB, DCN, RGN, KISS1, CHI3L1, SREBF1, IQGAP1, BSND, FGF7, ZNF35, VASH2, YY1, ESR1, AGT, PEAK1, TFF1, TFRC, NOTCH3, EZR, COX8A, FBP1, CLDN4, HAVCR2, VAV1, CRYZ, FASN, TIMP2, MUC6, TSPAN8, MYB, MIR196B, TXN, MIR330, ATF2, MIR31, IL33, AGTR1, VSIR, CES2, LOXL2, TNFRSF10C, PGK1, SSTR4, ABCC3, PEBP1, CEMIP, CLIC1, EGLN3, ADRA1A, PRDM14, HHIP, SLC52A2, MIR29B1, CDCP1, ADRA2B, MIR29B2, TAM, KLK7, MAP3K1, CCR7, ALCAM, MAP3K7, MGMT, MIF, CXCL10, EDNRA, NEK2, RASA1, TPX2, NEAT1, EEF1B2P2, PRMT1, GJB2, KDM1A, APLP2, CXCR6, RBM14-RBM4, MIR146A, GJA1, KANK2, NOX4, REG1A, USP22, MIR150, CA9, HTC2, MAP2K2, CD40, GGT1, GPR42, DKK1, MIR15B, E2F1, PDCD4, FAS, AREG, HHLA2, GOLGA6A, HPGDS, BHLHE22, HNRNPA2B1, MCAT, GOT1, SLCO1B3, RGCC, EIF4E, LILRB1, MIR106B, NRG1, PTK2, PHGDH, PWAR1, CCKAR, ARF6, GSTP1, MSH6, MARK2, EBI3, IGFBP1, S100A8, CDK1, S100A9, RBPJ, S100A11, CAT, GATA4, MIR191, SATB1, CDA, MIR1247, ENO1, CBX7, DNM1L, ABCC5, SERPINA1, SCO2, ARL2BP, BRS3, CHST15, CHEK2, GRP, AKR1A1, GSTM1, PLG, ECT2, PRNP, RPL10, SDF4, MIR17, RBM14, MIRLET7B, PLAC8, ADH1A, TRIM29, THY1, NAF1, C17orf97, ASPH, PTPN1, TCF7L2, TWF1, TPD52, PTPN11, TP53BP1, LPAR1, MTDH, TK1, LMLN, TLR1, TNS4, TGFBR1, TFF3, CRY2, CRMP1, AR, FAM83A, PTK6, PTBP1, TFAP2A, E2F3, KISS1R, TGFB2, MAK16, TIAM1, CCL21, TBX2, IDO2, CYP24A1, RFXAP, CASC2, ROBO1, DNM2, ROS1, SMARCA2, RBM45, RPS6KA2, RPS6KB1, RPS27A, ABHD11-AS1, S100A2, RELA, SIX1, TRIM69, SFRP4, ARHGAP4, LDLRAD3, SEL1L, SARDH, RMDN2, CCL2, PDIK1L, TICAM1, CCL18, DNMT3A, LRG1, TAT, DLX6-AS1, TACR1, PTPN14, TAC1, PTPRC, SOX2-OT, RAB5A, SYT1, STING1, GADL1, PRRT2, CDCA5, DUSP1, STK4, CYP2B6, STAT5B, SNHG15, CYP1A1, DPYSL3, DNMT3B, CYP1A2, CYP1B1, OSBPL5, SSAV1, RECQL, SRY, SRF, AZIN2, BHLHE23, TPM3, CD68, IL23A, PRMT3, PSME3, ABCC4, B3GALT5, GOLM1, TUBB3, ING4, PRMT5, WASF2, VAV3, RMDN1, SEMA4D, DESI2, ATG7, ANP32B, CD40LG, FOXP3, GDE1, ZFR, SLIT2, DLL4, MEG3, COX5A, RMDN3, CDH17, HEATR1, SLC52A1, CDH3, PAF1, IQSEC1, NAMPT, MFN2, KRT20, TLR9, BCL2L11, PTPRU, CDC25B, WWOX, CDC20, ASAP1, TXNIP, BTK, PRPF31, KLF8, DDR1, CTRC, BBC3, CILK1, MYOF, ZHX2, NOC2L, SMG1, CD36, SULF1, CASP9, TFIP11, SLC7A8, ANGPTL2, SNHG1, SRRM2, DAPK2, RASSF1, MAP4K1, FSTL1, SRPX2, DELEC1, CELF2, CTCF, CD86, ERO1A, SLC39A3, AP4B1, EEF2K, GIPC1, CD80, UHRF1, TBK1, MCTS1, CD14, SGSM3, RBMS3, WDR5, CDK8, CDK9, TPT1, COL4A2, COL11A1, WNT10B, DHDDS, XPC, XRCC3, XRCC4, BAX, ZFP36, COL1A2, ULBP2, LIN28A, MANF, MCPH1, TUSC3, TCTN1, BHLHE41, CMM, FOSL1, LAT2, VIP, SYBU, UBC, NR2C2, TRAF2, TRAF6, TMPRSS13, CPOX, TTK, ITCH, TYRO3, NUF2, BAG1, ARHGAP24, ATR, AVP, CLDN3, UTRN, KDM6A, UVRAG, VCL, USP5, LTB4R, WNK1, HAP1, KSR1, CDK5R1, PER3, PER2, PNO1, SQSTM1, BNC1, DIABLO, METTL3, USP9X, TM4SF5, ARTN, AIP, DMAP1, SLC16A4, SLC16A3, DYRK1B, BUB3, CES1, CCN5, CFL1, CHPT1, CMA1, GORASP1, ROBO3, CUL4B, IL21, CIP2A, DENR, CLU, DANCR, TP73-AS1, SMURF1, TNFSF13, AKR1B10, CEACAM1, TNFRSF10D, TNFRSF10A, IL18R1, CCR5, PADI4, MIR629, LINC01672, HSPD1, PAWR, HSPG2, PARN, ITGAV, C20orf181, MME, EPS8, ERBB4, FGF10, ITGA3, GFRA2, PRDX1, SNHG16, MIR451A, PAEP, AKR1B1, MMP11, ALDOA, P2RX7, P2RX5, ALDH1B1, GRK2, MIR296, OLR1, MNAT1, MAP3K10, ITGB2, NTS, MIR204, EPHB4, PFN1, MIR483, KCNJ11, KCNH2, MIR509-1, PER1, PDPK1, MIR497, HOXB7, MIR193B, MIR885, MIR373, HSPA4, ITK, HOXD13, GLI3, PDK1, FGFR4, MICOS10-NBL1, PDCD1, ANGPT1, MITF, TMX2-CTNND1, HSPA2, ANG, IFIT3, ROR2, PHB, TMEM238L, FBN2, MIR1231, IL6ST, IL17A, TNFRSF9, MIR135B, GAPDH, MIR1246, IL9, MTRR, FAT1, MIR148B, FAP, MIR424, MIR375, IL12A, CISD2, NEDD9, IL15, NEDD4, NCL, NBN, NBL1, MUTYH, IL13RA2, MIR374A, MTAP, IL2RA, ADAM8, NPY, NTRK2, NTF3, IRF2, FDPS, MIR1290, FCN2, FBL, GCHFR, PNP, IGF2R, ILK, MSMB, ESRRB, NGFR, NOS2, MIR1291, MSN, MIR1181, FCGR3B, MIR99B, AFP, IL18, NME1, MIR1271, FCGR3A, IKBKB, MIR744, MAP3K5, PRRX1, ELAVL1, POTEF, PPIA, GGT2, APOA2, FOXL1, EIF4EBP1, LASP1, RPSA, CCR2, EIF5A, CXCR3, FOXC1, GGTLC4P, MIR144, BIRC5, PODXL, HFE, UPK3B, HIC1, SNHG14, CDKN2B-AS1, PMS1, SMAD7, MIR15A, MR1, MIR139, LDHB, LTB, GSN, RELN, GRB7, PRSS2, AQP3, EEF1A2, MUC5B, CXCL1, GSK3A, FADS1, MIR601, MIR10A, PRKCB, MIR130B, MIR122, ANXA8L1, PRKAR1A, MIR126, GGTLC3, LRP2, GGTLC5P, ANXA8, EIF4A1, FOLR1, LTA, MAGEA3, CEACAM6, KCNN4, MCC, HNF4G, PCAT1, MCAM, GLO1, HNRNPK, KCNQ1, ENG, MIR196A1, MIR192, MIR185, PLAUR, SLC25A3, PLAT, MAP1A, LINC-ROR, MIR455, FKBP4, MIR181C, MIR183, MIR486-1, FOXA1, BMP8B, PIMREG, MTPAP, ZNF654, RHOF, NLRP2, CCL28, LTB4R2, MRGBP, PRPF40A, MIR874, ERO1B, MIR940, EIF5A2, STYK1, MIR646, MIR92B, IL17RB, LXN, MAML3, FERMT1, SMARCAD1, RIPK4, MIR652, TRPV6, ETNK1, TREM1, TERF2IP, KCMF1, SPHK2, BMP3, CHD7, ANLN, GPR137, MIR374B, MIR935, RETN, STK31, BNIP3L, GACAT2, ADPGK-AS1, OGDHL, CEP55, SNHG9, SARS2, CNDP2, OCIAD1, AGAP2-AS1, INTS11, MIR675, NKRF, ZGLP1, MSTO1, SLC35F6, LINC00462, MIR891B, MIR365A, BOK, PSENEN, ANO1, PIWIL2, KDM3A, PRR11, EXOC2, MIR876, CDKAL1, SULF2, SOX6, NSUN5, MIR1225, SLC38A7, BMP2, DIP, PARP14, KDM7A-DT, UGT1A1, MIR760, UGT1A3, PBRM1, ASPN, LOC729970, BPHL, CC2D1A, RADIL, ST7L, ALKBH5, HHAT, SLC25A36, SYCE1L, DST, MYDGF, MED29, KPNA7, PCBP3, GLS2, ERVK-11, MYB-AS1, LSM1, BANCR, LINC01111, HCAR1, GHET1, CA2, AGO2, SERPING1, LOC102723407, GATD3B, DKK4, SIK1B, CDR1-AS, CAD, CALCR, ANKRD1, SPINK4, MIR5100, UBE2S, RABGEF1, ATAD2, HIPK2, MACROD1, NOB1, ERVK-22, MIR2682, MIR4656, LAMTOR2, REM1, DLL1, SPRY4-IT1, SLCO3A1, SLCO4A1, HTRA2, DESI1, PSC, C3, DNAI1, USP21, SNORA74A, LETMD1, LOC112694756, MOB4, ZEBTR, PRPF40B, LNCRNA-ATB, EML2, CADM1, RAB38, SH3BP4, TMEFF2, ACADS, H3P13, H3P8, ABCA4, FAM215A, DDX58, SERPINA3, USP49, CASP7, IRAIN, LOC110806263, EHF, PERCC1, FGF21, PTPN22, CERNA3, KCNK15-AS1, LINC00976, CAPN1, NUTF2P3, CHD5, SIRT1-AS, ACO1, NECTIN3, CAST, CLIC4, MYRIP, TMEM158, C1GALT1C1, LINC00339, DUOX1, ADAM10, PRKAG2, CRLF3, ZMYND10, MIR1301, MIR1180, MIR634, TLR8, BTC, KLF9, C1QBP, CLDN18, CKLF, ZEB2-AS1, HSPA14, LEF1, MIR663B, MIR4306, ACVR2A, YTHDF2, SIRT6, RAB14, NT5DC3, GPR87, LOXL1-AS1, MBD3, IL17D, LINC00994, BCL11A, ADA2, MAP3K20, CDK12, CMPK1, CAB39, RAB23, SF3B6, MIR1297, PIAS4, MIR1243, MIR1266, MIR3686, A4GNT, INSIG2, IL20, DUOX2, ERVW-1, ACTA2, VSX1, KLRC4-KLRK1, ACP3, UBQLN1, BUB1B, ERVK-2, STRN4, NPC1L1, MIR3064, TSPO, BRD7, MYLIP, ERVK-12, PYCARD, ACVR1, IL21R, IRAK4, PXN-AS1, ANGPTL4, MIR642B, MIR3923, APIP, GMNN, MIR3679, ISOC1, LINC01133, BTF3, ASCC1, SMIM31, TRAT1, LINC00958, F11R, TAS2R10, LINC01006, ARHGEF4, SLC25A37, DIO3OS, EMSY, PROKR2, CD109, ADAT2, GPR151, SLCO6A1, EMB, REG3G, ANPEP, MIR208A, ANGPT2, FATE1, MIR211, TMIGD2, SPNS2, MSI2, ARR3, JDP2, SPPL3, H4-16, CCDC26, MUC17, GATA5, ANXA5, LINC00052, MIR198, MIR199A1, PTGR2, PRIMA1, FBXL14, MIR199A2, MIR19A, MIR19B1, SYT9, MIR20A, CACUL1, ANXA11, ANXA6, TMEM37, BRI3BP, SLC32A1, CYP2R1, TMEM45B, PRAP1, MAL2, MCM3AP-AS1, ALDH1A3, IL17F, MIR28, EGLN2, AGTR2, PPP1R14A, MIR302A, ART1, FOXQ1, ASAH1, DMKN, ASNS, MIR30C1, MARVELD3, LRP5L, MIR30C2, NLRP3, MIR155HG, ALPG, MIR224, MIR215, TWIST2, MRGPRX4, MRGPRX3, DCD, ANIB1, TM4SF18, CMTM5, CTHRC1, MIR219A1, NOSTRIN, MIR22, UHRF2, ALOX15, FBXO32, ALK, CSMD2, MIR197, SIK1, APAF1, MIR127, MCIDAS, MIR129-2, RSPO2, KLK3, ANO9, APRT, CHSY3, TRIM59, APOC1, ARG1, LYPD5, LINC00346, LINC01559, OR10A4, BCL9L, APOA1, SLC25A45, MACC1, PWAR4, SBF2-AS1, DNAAF3, CCSER1, PAIP2B, LINC01121, C16orf74, HRNR, MIRLET7C, SERPINA13P, CEP85L, MIRLET7D, LINC-PINT, MIR100, MIR106A, FASLG, ENTPD8, GSTK1, TMPRSS9, MIR125A, H19, ASPM, MIR193A, MUC20, ARID2, B3GNT6, MIR182, ADAMTS18, OIT3, BHLHA15, OXER1, IFNLR1, SLFN5, EML5, CLEC14A, MIR186, SLC5A8, RHOB, FEZF1-AS1, NKAIN2, DAB2IP, MIR181A2, CENPX, MRGPRX1, MED19, MIR149, SGMS1, NPCA1, FAM133B, ARG2, MIR152, RICTOR, IL27, SEMA3D, BIRC2, GPRC6A, FOXK1, KDM1B, MIR18A, ALKBH3, MARCHF8, VWCE, LINC00473, MIR32, SMYD2, SMURF2, NIBAN2, YTHDC2, PORCN, IL25, DEPTOR, ILRUN, S100PBP, SMYD3, TMPRSS3, PNPT1, HIF3A, COP1, RHBDF1, MMS19, LINC01060, BGLAP, MIIP, SAV1, MIR520H, PINK1, MIR519D, MIR181D, BCL6, DHX40, DYNC2H1, TMEM204, MIR489, MIR146B, BCR, MIR202, PLEKHF1, MIR492, BDNF, MIR493, CFB, GGCT, STK33, ADRB3, CDK15, AFAP1, LGR6, ACE2, SLURP1, ADH4, PNPLA2, CXADRP1, CIAPIN1, ADAMTS9, PRDM1, H3P38, MIR539, SNORA25, MIR421, MIR584, KIF15, OTUD7B, MIR607, MIR608, MIR613, MIR623, CD248, NORAD, IL22RA1, ADH6, SNX6, SINHCAF, HAMP, ANKRD36B, KLHL1, MRTFA, CFAP97, PCDH10, CERNA2, CRNDE, MIB1, RPTOR, ARID1B, PPM1H, BGN, AICDA, PARP4, MIR485, MIR410, BCKDHB, VN1R17P, EBPL, GPR166P, FOXB2, XRCC6P5, ZGPAT, NT5C1A, MCHR2, HOPX, MCM8, RAB11FIP4, PROK1, RPAIN, MIR331, AP2A1, MIR340, USP44, MIR367, TRIM63, KDM2B, MIR370, MIAT, TSLP, NKD1, MIR320A, MIR34B, SNHG7, MIR93, JAG1, RAB2B, ATAD1, PARP10, MIR99A, RIOX2, KMT5C, PSRC1, CRISP1, ASTL, HINT2, MYCBPAP, ATF3, SH3D21, PPP1R2C, GRHL2, DUXAP8, JADE1, DUXAP10, NANOG, UBA5, NBDY, MFSD13A, KDM8, MIR448, MIR449A, H4C15, MIR431, MMRN2, MYH14, BCHE, MIR452, NAA25, CEP70, MIR371A, MIR425, RERE, IMMP2L, HASPIN, SUMO1P3, SESN2, VANGL1, NETO2, MIR377, MIR381, MIR384, NECTIN4, AZGP1, ZFP91, ITIH5, SLC44A4, SRCIN1, BAK1, GPX2, HS3ST3B1, LPAR3, PLOD2, EFNB2, PKN1, EGR1, PRKAR2A, PRKACG, EGR3, EIF4A2, PRELP, PPP5C, PPP1R1A, PPID, PPIB, MED1, PPARA, PON2, PON1, POLE, POLD1, PNMT, PNLIP, PMS2, PMS2P3, PMAIP1, PRKCZ, PRKDC, EFNA2, PTAFR, PTPRJ, PTPRG, PTPRA, PTPN13, QSOX1, PTN, E2F2, PTGS1, PTH, PTGER4, PSMD10, EEF1G, PYY, PRTN3, KLK10, HTRA1, KLK6, PRSS3, EDNRB, PROX1, EEF1A1, MAPK9, PLXNA1, CELA1, DYRK1A, PLEK, CDK16, PCBP1, PCBD1, PBX3, PAX6, PAX5, PAK3, PAK2, EREG, SERPINB2, ERCC4, PAFAH1B2, PCSK6, P2RY6, P2RY2, OSM, OSBP, ORM1, OPRM1, OPRD1, TNFRSF11B, ERCC5, ODC1, PCYT1A, PDE8A, PDGFA, PIGR, ELK3, PLCB3, EMD, PLA2G4A, PKLR, PKD1, PIN1, EPHA1, EPHA4, EPHB1, EPHB3, PDGFB, PGR, PFKP, PFKM, PFKFB4, PFDN5, PEG3, PECAM1, PDZK1, ENPP1, PDK4, PVR, RGL2, PSD4, SAFB, SLC1A2, DCX, SKIL, SKI, SIM2, ST3GAL4, ST6GAL1, DDB2, SFRP1, DDT, SELP, CX3CL1, XCL1, CXCL5, DLD, CCL8, CCL7, DMD, SRL, SCTR, SCN10A, SCN9A, SARS1, ACE, SLC5A1, DCTD, DAG1, SPP2, CYP2C19, SPG7, CYP3A4, SOX12, SOX15, CYP51A1, SOS1, SOD3, DACH1, SNCG, SLC6A8, DAP, SMO, SMN2, SMN1, SMARCC1, DAPK1, SLPI, DAPK3, SLC9A1, SLC8A1, SAI1, S100B, RAB27A, S100A10, RGS16, TRIM27, REST, REN, RDX, RBP4, RBP1, RBM3, KDM5A, DPEP1, RASGRF1, PLAAT4, RARB, RAP1B, RAP1A, RALA, RAG2, DRD2, HBEGF, RAD17, DVL2, RABIF, RAB27B, RNASE1, RNASE3, RNASEL, RPS8, S100A7, S100A6, DNA2, SORT1, S100A1, RXRA, DNAH5, DNASE1, DYNC1H1, RPS15A, RPS6KA3, RNF2, RPS6KA1, RPL39, RPL34, RPL29, RPL26, RPE65, RORB, RORA, SNORA73A, BRD2, ERG, NTRK3, ERN1, IARS1, GAS6, GATA1, GATA2, GATA3, IGSF1, GC, IGFBP7, IGFBP2, GCK, GCKR, GCNT1, IFNB1, IFNAR1, IFNA17, GCY, GDF2, GDF10, IFN1@, GFPT1, IDH1, ID2, IRF8, ICAM2, CXCR1, GALNT3, GAD2, ITGB4, KRT8, FRZB, FUS, KCNK2, FUT3, FUT4, GABBR1, JAK1, JAG2, EIF6, GABRA3, GAD1, ITGA5, ITGA1, ISG20, IRS1, IRF1, ITGA6, GABRP, INSR, INPP4A, IDO1, GFRA1, HYAL1, NT5E, HTR1D, HLA-DRB1, HLA-DOA, HLA-C, HLA-B, GOT2, GP2, HGFAC, GPI, HDGF, HCRTR1, HCRT, H2AX, H1-5, H1-4, GUSB, GUCY2C, GPT, GSTM3, GPX1, GRM1, GRIA3, GRB10, GRB2, GNRHR, HMGB2, HMGCR, HPD, HTR1B, HSPE1, GHR, GHRH, HSPA9, GJB1, HES1, AGFG1, GPC3, HPGD, HOXB9, GLUL, HOXB8, HOXB6, HOXB2, HOXA1, HNRNPU, HNRNPC, ONECUT1, HNF4A, GLUD1, HMMR, KRT17, KRT18, KRT19, FPR2, NEDD8, MYH9, MYD88, FANCD2, MMUT, TRIM37, PTK2B, MTR, ND2, FBLN2, MT1M, MRE11, MPST, MPO, MPL, MMP16, MMP15, GPC4, MMP13, MMP12, FER, MMP3, GPC5, NELL1, NEO1, NEU2, NQO2, YBX1, NRTN, NPY2R, NPM1, NPC1, CCN3, NOTCH2, NOS1, NOP2, NNMT, NM, NEUROD1, NHS, ETV4, NFYA, ETV5, EWSR1, EYA2, NFE2, F9, F10, NEUROG1, KMT2A, MAP3K9, MKI67, FLNA, LMO2, LMNB1, LLGL1, LIPE, LIMS1, LIMK2, LIMK1, LIG4, LIG3, LIFR, FLNB, LPA, LGALS3BP, FOLH1, LFNG, LEPR, FOLR2, LBR, LAMC2, LAMP1, LAMB3, LAG3, LOX, LRP1, MICB, VEGFD, MFAP1, FH, MAP3K4, MAP3K3, MEF2D, MEF2A, MECP2, ME2, FHL2, MDH1, MATK, LRP6, MAT1A, MAP2, SMAD6, SMAD3, SMAD1, FOXD1, TACSTD2, FLI1, BCAM, LTA4H, SPOCK1, CYP2B7P, SREBF2, MVP, GDF11, TRIM13, RBM5, RBM6, TRIM28, G3BP1, ARFRP1, FRY, LRPPRC, RAD50, PPIF, ACTR2, ARPC4, EDIL3, IL18BP, ABCB6, GNE, HDAC6, AKT3, HNRNPDL, NUP153, NR1H4, NR1I3, TRIB1, CNKSR1, SF3B4, CD74, DDX17, CD63, SEMA3C, CARM1, CAP1, MERTK, TACC3, GPNMB, BASP1, RACK1, DLC1, CDK2AP2, IRF9, TUBA1B, MICU1, KLF2, WARS2, TLR6, TNIP1, RTN3, SF3A1, STUB1, MED12, USP15, MSC, GPX4, LITAF, MAGED1, ROCK2, ATG5, RASAL2, MAP4K4, AIM2, GSTO1, QKI, NTN1, RECQL5, CD101, KIF3B, SLC9A3R1, SLC9A3R2, CD163, SOCS6, SNORD22, ADGRG1, BCL7B, PIWIL1, MAPKAPK2, AIMP1, SCAMP1, BAG5, CDH13, SDC3, DLEC1, KIF14, NUAK1, TLK1, CDC25C, MTSS1, SETD1A, USP34, CDH5, DCAF1, MDC1, PTGES, SOCS5, TTLL4, WTAP, PRDX6, APOBEC3B, SPAG6, CDH11, CHD1L, CXCL14, EI24, NEBL, CD59, ARPC1A, HTATIP2, KLRK1, ARHGEF15, MLXIP, FASTKD2, NLRP1, VASH1, CCNA2, ITGA11, CBX3, TUSC2, PDAP1, PHB2, GPR182, SLCO2B1, KLF12, FILIP1L, SLC6A14, GALNT5, CCNB1, WIF1, MAP4K5, RPP14, ADAMTS8, KIFAP3, SIRT2, STK38L, NCAPH, ZNF281, PES1, ICMT, CELA3B, GPR161, TPSD1, CAV2, RUNX1, SIRT4, DICER1, NCSTN, CBR1, SATB2, SIN3B, BICD2, MPRIP, CBL, KDM6B, PEG10, KDM4C, PDS5B, TNIK, RER1, TRIM31, TOPBP1, KHDRBS1, PLK2, KDM5B, RBBP9, PLK4, NFAT5, POLQ, USP39, CCT8, DLL3, TNFSF13B, SPINT2, WASF3, SCGB1D2, IGF2BP2, CD34, IGF2BP1, ENTPD1, PDLIM5, SLCO1B1, ENTPD2, SMC2, OLFM4, SEPTIN9, FRS2, UBE2C, STIP1, OGFR, RIPK3, DSTN, RAB31, CCND2, CCND3, KCNQ1OT1, COPS6, FERMT2, CKAP4, CCNG1, CD28, EHMT2, PAPOLA, GADD45G, BRD8, MALT1, PPARGC1A, MRPS30, NMU, ME3, ADAM28, GCNT3, CDKN1C, SRPK2, TNFAIP1, COMP, COMT, SLC31A1, CPE, NR1H2, UCK2, UGT1A, UGP2, UCHL3, UCHL1, UBE2N, CPN1, TULP3, TUFT1, TTR, TRH, HSP90B1, TPO, TPD52L2, CREBBP, CREM, TOP1, TNFRSF1B, BEST1, TRPV1, VTN, XRCC2, NELFE, DEK, SCLC1, PAX8, BTG2, ZNF154, ZBTB16, CNBP, ZFX, YES1, COL5A2, WARS1, COL6A1, XPA, XK, COL6A2, COL6A3, WNT7B, WNT2, WNT1, COL17A1, WAS, TNFRSF1A, CRKL, PTTG1, CLDN5, CTPS1, HNF1B, CTRL, TBX3, TCEA3, TCEA2, TCEA1, CTSB, TAGLN, ADAM17, CTSD, SULT1A1, CTSE, CTSH, STIM1, CX3CR1, STAT2, CXADR, ST13, CYP2A6, CYP2A13, SSR2, TRIM21, CST1, PPP1R11, TDG, CS, TMSB4X, TMPRSS2, TM7SF2, TLR3, TLR2, TLE1, CRY1, THBS2, THBD, THAS, TGIF1, PRDX2, CSF1R, TGFB1I1, CSF3, CSN2, TFF2, TFCP2, TF, CSNK2A2, TERC, VCAN, TFEB, COL1A1, CNC2, ABCC2, SPHK1, ARHGEF7, IER3, ASAP2, DLEU2, CCN6, NRP2, INPP4B, CHGA, RAB11A, CHKA, ADAM15, CHRM3, CHRNA7, TNK1, BECN1, EIF3F, IRS2, DYNLL1, CLCA1, NUMB, PTCH2, RUVBL1, FUBP1, NAE1, MBD2, ATP6V0D1, MTA2, AURKB, CDKN2D, ARHGEF2, SCAF11, TMSB10, EBAG9, EXO1, ATG12, CCNE2, CDO1, WASF1, DIRAS3, CLDN8, CDX1, BRSK2, CLIC3, MAP3K14, F2RL3, CEBPB, P4HA2, CDK5R2, NOP14, AKR7A2, MADD, UBL4A, H4C11, H4C12, H4C6, H4C4, H4C1, FZD6, FZD4, CDC7, H4C9, ARID1A, GATD3A, H4C8, SLC7A5, ANP32A, GPR68, YEATS4, KMT2D, MFAP5, RASSF7, ADAM12, SLC25A16, CUBN, H4C3, H4C2, LMO4, TTF2, HAT1, PARG, PIK3R3, PPFIA4, CLTC, GEMIN2, SEMA7A, OGT, SORBS2, KLF11, DOC2A, H4C5, RAD54L, STK24, TAGLN2, SPARCL1, PIP4K2B, PIP5K1B, PIP5K1A, NME5, H4C14, H4C13, H3P40
-
Quartan Fever
Wikipedia
Spraying is a method in multiple regions and to control epidemics. [10] Nets treated with insecticide are effective in preventing mosquito contact for three years. ... House improvements including windows, installation and sealed doors reduce the risk of coming in contact with the infected mosquitoes. [10] Larval source management is the control and monitoring of aquatic environments in order to prevent fully Anopheles mosquitoes from fully developing. [10] Mosquitoes require aquatic environments in order to fully mature and develop. ... When removing any water-filled containers from the surrounding area the mosquito life cycle is halted and acts as a method to reduce mosquito population within the surrounding area. [11] Clothing can act as a physical barrier to prevent exposure of flesh for mosquitoes to feed on, treating beds and clothing with insecticides/repellents can further reduce chances of infected mosquitoes from biting and passing quartan fever to individuals. [10] Avoiding areas which have high mosquito populations, specifically for quartan fever the P. malariae strain. [10] Avoiding travelling to regions which have a sub-tropic climate to prevent infection and developing quartan fever. [10] Implementing the sugar baiting method aids in reducing the population of Anopheles mosquitoes, and ultimately reducing the likelihood of catching quartan fever. ... DAILY MED . 8 July 2010 . Retrieved 10 May 2019 . ^ a b "Malaria" . 2019 . Retrieved 10 May 2019 . ^ a b Kayentao, K (23 February 2012).
-
Clanging
Wikipedia
This is associated with the irregular thinking apparent in psychotic mental illnesses (e.g. mania and schizophrenia ). [1] Gustav Aschaffenburg found that manic individuals generated these "clang-associations" roughly 10–50 times more than non-manic individuals. [2] Aschaffenburg also found that the frequency of these associations increased for all individuals as they became more fatigued. [3] Clanging refers specifically to behavior that is situationally inappropriate. ... Livingstone. p. 32 . ^ Spitzer, Manfred (1999). The mind within the net: Models of learning, thinking, and acting .
-
Histiocytosis
Wikipedia
According to the Histiocytosis Association of America , 1 in 200,000 children in the United States are born with histiocytosis each year. [2] HAA also states that most of the people diagnosed with histiocytosis are children under the age of 10, although the disease can afflict adults. ... Information concerning histiocytosis and clinicians located in European countries may be found in many languages at the web portal of Euro Histio Net (EHN). This is a project funded by the European Union, coordinated by Jean Donadieu, APHP , Paris, France. ... The Society has instituted several clinical trials and treatment plans. [9] [10] References [ edit ] ^ Histiocytosis Archived 2016-10-09 at the Wayback Machine at eMedicine Dictionary ^ Disease information at the Histiocytosis Association of America ^ "Histiocytosis - Signs and Symptoms" . ... Report of the Clinical Advisory Committee meeting, Airlie House, Virginia, November, 1997" . Ann Oncol . 10 (12): 1419–32. doi : 10.1023/A:1008375931236 . ... External links [ edit ] Classification D ICD - 10 : C96.1 , D76.0 ICD - 9-CM : 202.3 , 277.89 MeSH : D015614 SNOMED CT : 60657004 External resources MedlinePlus : 000068 eMedicine : ped/1997 v t e Histiocytosis WHO-I/ Langerhans cell histiocytosis / X-type histiocytosis Letterer–Siwe disease Hand–Schüller–Christian disease Eosinophilic granuloma Congenital self-healing reticulohistiocytosis WHO-II/ non-Langerhans cell histiocytosis / Non-X histiocytosis Juvenile xanthogranuloma Hemophagocytic lymphohistiocytosis Erdheim-Chester disease Niemann–Pick disease Sea-blue histiocyte Benign cephalic histiocytosis Generalized eruptive histiocytoma Xanthoma disseminatum Progressive nodular histiocytosis Papular xanthoma Hereditary progressive mucinous histiocytosis Reticulohistiocytosis ( Multicentric reticulohistiocytosis , Reticulohistiocytoma ) Indeterminate cell histiocytosis WHO-III/ malignant histiocytosis Histiocytic sarcoma Langerhans cell sarcoma Interdigitating dendritic cell sarcoma Follicular dendritic cell sarcoma Ungrouped Rosai–Dorfman disease
-
Malaria
Wikipedia
The mosquitoes remain on the wall until they fall down dead on the floor. Insecticide treated nets [ edit ] A mosquito net in use. Mosquito nets help keep mosquitoes away from people and reduce infection rates and transmission of malaria. Nets are not a perfect barrier and are often treated with an insecticide designed to kill the mosquito before it has time to find a way past the net. Insecticide-treated nets are estimated to be twice as effective as untreated nets and offer greater than 70% protection compared with no net. [73] Between 2000 and 2008, the use of ITNs saved the lives of an estimated 250,000 infants in Sub-Saharan Africa. [74] About 13% of households in Sub-Saharan countries owned ITNs in 2007 [75] and 31% of African households were estimated to own at least one ITN in 2008. ... That number increased to 20.3 million (18.5%) African children using ITNs in 2007, leaving 89.6 million children unprotected [76] and to 68% African children using mosquito nets in 2015. [77] Most nets are impregnated with pyrethroids , a class of insecticides with low toxicity . ... The other species usually cause only febrile disease. [132] Severe and complicated malaria cases are medical emergencies since mortality rates are high (10% to 50%). [133] Recommended treatment for severe malaria is the intravenous use of antimalarial drugs.ICAM1, FCGR2B, HBB, CD36, NOS2, FCGR2A, TNF, CR1, G6PD, CRP, HP, ACKR1, GYPA, SLC4A1, GYPB, NCR3, TIRAP, GYPC, LTBR, CISH, IFNG, HMOX1, PKLR, ABO, ANK1, AQP4, ATP2B4, HBG2, CYTB, ENOSF1, MSMB, MST1, ZNF536, LINC00944, SMARCB1, DHODH, PDR, TREML4, ZNF804A, OR51F1, OR51B5, CDH13, PROCR, SPATA3, OR51N1P, DHFR, DDT, RECQL4, FAM155A, IGHG3, IL4, MMP26, IL6, IL10, TLR9, HLA-DRB1, CSMD1, HBE1, DNAJC5, TMPRSS13, KLHL3, HDGFL2, TLR4, ATAD1, LMLN, TENM3-AS1, MECP2, POMGNT2, MBL2, TFRC, TGFB1, MIF, HLA-B, HAMP, DHPS, SERPINA3, TLR2, IL1B, FOXP3, FHL5, ACOT7, POTEKP, POTEM, GEM, KIR3DL1, RN7SL263P, ACTG2, ACTG1, ACTB, ACTBL2, HBA2, CYP2B6, HSPA4, LSAMP, TRAP, FCGR3B, HSP90AA1, IL1A, LAMP3, CD81, OR10A4, CCL5, ABCB1, FAS, CD40LG, TEP1, CXCL8, IARS1, HLA-G, CTLA4, HBA1, INSRR, ANGPT2, TYMS, CFH, GSTP1, IFNAR1, AGT, GYPE, FCGR3A, TXN, IL13, HSPB3, APOE, MTCO2P12, ISYNA1, FCGR2C, FYB1, VDR, HLA-A, GSTM1, GSR, ATR, MBL3P, LAIR1, PNP, IL12B, MNAT1, IL1RN, CYP2D6, IGF1, CD55, ACHE, DECR1, COX2, IL3, CCL2, MAPK1, NLRP3, FBXW7, HAVCR2, THBD, VPS51, EMP1, ITGA2B, PTGS2, ANC, IL10RA, XPO1, VNN1, PLEK, UMPS, IL2, IL2RA, TPPP, VWF, ISG20, ADAMTS13, IRF1, IL7R, AIMP2, IL12RB1, CLEC11A, METAP2, CDK5R1, ING1, IL18R1, PGD, HAP1, H6PD, PRDX5, GRAP2, CXCL9, MMP9, MPO, TAP1, CCL4L2, COX1, EBI3, ITGAX, COX3, TLR6, CXCL11, MTHFR, NFKB2, NFYA, NOS1, TBC1D9, ORC1, MCF2, AKAP13, RNF19A, TLR7, NT5C3A, IRAK4, KIR2DS1, CCL4, KIR3DL2, ICOS, COQ2, PSIP1, PECAM1, TPT1, RNASE3, ARTN, TP53, POLDIP2, PDCD1, TLR1, AHSA1, UBL4A, AQP3, AGRP, H3C9P, CYP2C8, CYP2C19, GTF2H4, CRK, RNA18SN5, ANXA2, H3P37, CASP1, NANP, CCL4L1, MAPK14, CXCR3, GNAS, GLO1, FCN2, SMIM10L2B, FKBP4, CD27, FOXO3, RBM45, HM13, IL33, HK1, CCR5, IFNA13, IFNA1, H3P42, DNAJB1, CHIT1, CYP3A4, SMIM10L2A, EGF, CHI3L1, CAT, EPHA2, NSFL1C, ADRB2, MYMX, COX8A, GAPDH, ABCB6, NR1I3, TREML1, PUM3, FMN1, TICAM2, TRIM13, BMS1, FZD4, RABEPK, LANCL1, FUT9, TNFSF13B, DCTN6, CXCR6, ARL6IP5, MRGPRX1, ZNRD2, ASPM, KAT5, RAB7B, CIB1, SEMA3C, ARMH1, STING1, CFDP1, CPQ, MYLK4, DLC1, AKR1A1, PIEZO1, TMPRSS11D, HDAC9, CARTPT, DEFB4B, TIMELESS, SPHK1, TMED7-TICAM2, PSC, VNN2, PROM1, UPK3B, H3P23, H3P28, TNFRSF11A, TNFRSF18, TP63, PDXK, CNTNAP1, DHX16, STK24, H3P19, LOH19CR1, WASHC1, WASH6P, LPAR2, MIR146A, APOBEC3B, SPAG6, CLOCK, ATG5, MIR142, AIM2, ABCG2, PCSK9, MIR155, NCF1, PPIG, MIR29A, VN1R17P, GPR166P, CD163, MIR451A, CXADRP1, ARHGEF2, CERS1, SPINK5, MASP2, GEMIN4, ACD, TLR8, MPPE1, MCPH1, HSPA14, RNF34, TMED7, ARMC9, PPP1R2C, IL22, TRAF3IP2, A1CF, PDCD1LG2, SLC44A4, SGSM3, MCAT, HPGDS, B3GAT1, ROPN1L, PHGDH, RAB14, IL23A, ABCG4, IFIH1, CFC1, BTNL2, MARCHF1, POLE4, CMC2, TMED9, ACKR3, PDXP, RHOF, AICDA, POLD4, RBM25, TOLLIP, TREM1, LGR6, ADA2, BACH2, ERAP1, GOLPH3, PARS2, KRT88P, TRIM5, IL17RE, CHP1, GPR151, NRSN1, EIF5AL1, CD160, APCDD1, ERFE, OXER1, DNAJB1P1, DSTN, GPRC6A, CCNI, ADIRF, EBNA1BP2, TMED2, EHD1, RNPS1, HPSE, SEPTIN9, SCLT1, NT5C2, SLC25A21, LEO1, NLRP12, TIMD4, CDCA5, DBA2, CARD16, PTPMT1, CGAS, RAB39B, TADA1, MRGPRX3, MRGPRX4, PGLS, PANX1, SPO11, LPAR3, CBX5, POFUT2, SPPL3, NBEAL2, LUC7L, PTPRC, FGF23, EIF5, FLT3LG, FLT1, FECH, FBN2, FBN1, FANCD2, F3, EPO, ENO2, ADGRE1, ELK4, ELF4, EIF5A, EIF4G2, CXADR, EGR3, EDNRA, EDN1, S1PR3, RCAN1, ATN1, DNMT1, DEFB4A, DHX9, ACE, DBP, CYP1A2, CYC1, GABPA, GCHFR, GDF1, GPR42, IL4R, IL1R1, IGFBP1, IFNGR1, IFNB1, IFNA2, IFI27, IDE, HTN3, HSPA9, HSD11B1, HRES1, HPRT1, HPR, HPGD, HMGB1, HLA-DOA, UBE2K, HGF, SERPIND1, HBG1, GTF3A, GSTT1, GSN, GPX1, GPT, GRK5, CYBB, CTSL, IL9, ANXA1, C3, BSG, BRS3, BRCA2, PRDM1, BCL2, BAX, ASPA, ASIP, ARR3, NUDT2, ANXA7, ANXA4, ANPEP, CSH2, AMBP, ALOX5, ALB, AHR, AFP, ADSL, ADRA2B, ADRA1A, ADORA2A, ADH1B, ADA, ACP1, ACACA, CAST, CASR, CD1B, CD1C, CSH1, CSF1R, CSF1, CS, CRYZ, CREM, CR2, CLDN4, CPB1, CNTF, CCR4, CLU, ERCC8, CTSC, CEL, CDC25C, CD69, CD68, CD40, ENTPD1, CD34, CD28, CD19, CD14, CD9, CD1E, CD1D, IL5, IL12A, FOSL1, SELE, SPTA1, SPP1, SPINK1, SPG7, SOD3, SOD1, SMN1, SLC16A1, SLC11A1, SLC6A7, SLC2A1, SGCG, SET, SEA, ABCA1, SDC1, CXCL5, CCL22, CCL18, CCL3L1, CCL3, CCL1, SAFB, SORT1, RPS19, RBP2, RANBP2, PEX19, SSR2, SSTR4, DENND2B, STAT6, DDX39B, PRRC2A, PFBI, RAB7A, CXCR4, MOGS, ZBTB16, TRPV1, VCP, USP1, TYRP1, TTR, TTPA, TRPC1, TRP-AGG2-5, TPO, TPH1, TNFRSF1B, TLR3, TGFB2, TRBV20OR9-2, TCN2, HNF1A, TADA2A, ADAM17, TAC1, STK3, PTPRH, PTHLH, IL15, KIR3DS1, MAL, MAF, LTB, LTA, LMAN1, LEPR, LDLR, LCN2, LBR, RPSA, LAG3, KRT13, KNG1, KIR2DS5, PSMD9, KIR2DL3, KIR2DL2, KDR, KCNG1, KARS1, ITPA, ITGB2, ITGAM, ITGAL, CXCL10, IDO1, ILF3, IL18, MAP2, MAP6, MEFV, MVD, PSMD7, PSMD2, PSMB9, PSEN1, PSAP, PRSS1, PROC, MAP2K1, PRKG1, PRKAR1A, PPP1R1A, PPARG, SEPTIN4, PLP1, PGM1, PGAM1, P2RX7, SLC22A18, TNFRSF11B, OMD, ODC1, NOS3, NQO2, NFE2L2, NEK2, MYD88, MYC, H3P5
-
Esophageal Food Bolus Obstruction
Wikipedia
An increasingly commonly recognized cause for esophageal food bolus obstruction is eosinophilic esophagitis , which is an inflammatory disorder of the mucosa of the esophagus, of unknown cause. [7] [8] Many alterations caused by eosinophilic esophagitis can predispose to food boluses; these include the presence of multiple rings and narrowing of the lumen. [9] When considering esophageal dilation to treat a patient with food bolus obstruction, care must be made to look for features of eosinophilic esophagitis, as these patients are at a higher risk of dilation-associated complications. [10] Other conditions that predispose to food bolus obstructions are esophageal webs , tracheoesophageal fistula / esophageal atresia (TOF/OA) and peptic strictures . [7] Food boluses are common in the course of illness in patients with esophageal cancer but are more difficult to treat as endoscopy to push the bolus is less safe. ... ] and the use of large-bore tubes inserted into the esophagus to forcefully lavage it. [17] [ unreliable medical source? ] Endoscopic [ edit ] The Roth net can be inserted through the endoscope to remove pieces of the obstructed food. ... Traditional endoscopic techniques involved the use of an overtube, a plastic tube inserted into the esophagus prior to the removal of the food bolus, in order to reduce the risk of aspiration into the lungs at the time of endoscopy. [7] However, the "push technique", which involves insufflating air into the esophagus, and gently pushing the bolus toward the stomach instead, has emerged as a common and safe way of removing the obstruction. [7] [18] Other tools may be used to remove food boluses. The Roth Net is a mesh net that can be inserted through the endoscope, and opened and closed from the outside; it can be used to retrieve pieces of obstructed food. ... Clinical Gastroenterology and Hepatology . 5 (10): 1149–53. doi : 10.1016/j.cgh.2007.05.017 . ... "Review of food bolus management" . Can. J. Gastroenterol . 22 (10): 805–8. doi : 10.1155/2008/682082 .
-
Childhood Arthritis
Wikipedia
Retrieved March 20, 2012, from Centers for Disease Control and Prevention: https://www.cdc.gov/arthritis/basics/childhood.htm ^ "Juvenile Arthritis" . www.rheumatology.org . Retrieved 2020-10-26 . ^ "Juvenile Arthritis" . www.arthritis.org . Retrieved 2020-10-26 . ^ a b c d e f g h i j k l m Arthritis Foundation. (2012). ... Retrieved March 20, 2012, from American College of Rheumatology: http://www.rheumatology.org/practice/clinical/patients/diseases_and_conditions/juvenilearthritis.asp ^ "Juvenile Arthritis" . www.rheumatology.org . Retrieved 2020-10-29 . ^ "Autoimmune Diseases" . medlineplus.gov . Retrieved 2020-10-29 . ^ Takken, Tim; van der Net, Janjaap J; Helders, Paul PJM (2001-10-23). ... External links [ edit ] Classification D ICD - 10 : M08 ICD - 9-CM : 714.30 MeSH : D001171 National Institute of Arthritis and Musculoskeletal and Skin Diseases - US National Institute of Arthritis and Musculoskeletal and Skin DiseasesSMAD3, JMJD1C, ADCY7, IL1RN, SLC11A1, CTLA4, LPP, NDEL1, BTG3, CD14, CXCL1, CXCR4, OSM, PLAUR, PLOD2, NRG1, ASAP1, MAPK1, SLC22A16, PGM5, RNF103, PROS1, TUBB2A, H2AC8, FOXP1, MIR22HG, SIPA1L1, TNFAIP8, ANKRD9, MAFF, KAT6B, GP1BB, GNG11, GMPR, BEND2, STAB1, FOSL2, FOSB, FOS, PER1, TNFRSF10C, AOPEP, H2BC8, HERPUD2, KRAS, MTSS1, KCNJ15, JUN, ITGB5, ITGA2B, ETNK1, FAM20A, MAP2, MAD1L1, RBM47, CXCR2, DYSF, HSPA6, WWOX, SH3BGRL2, MSN, NR4A3, MS4A4A, MEX3C, IGF1R, NR4A2, C2orf88, MAML2, ACRBP, OLR1, FCGR1A, AVL9, TTLL5, CMTM2, CALD1, BTG1, RHOBTB1, TFPI, LILRA5, C8B, TFDP1, TREML1, CREM, PACSIN2, CD8B, TCF7L2, STIM1, CLU, STAT3, CD83, THBS1, FAXDC2, RERE, KLF4, B4GALT5, AREG, FCHSD2, AQP9, PCYT1B, TNFAIP6, UBE3C, ALOX12, MYZAP, WASF3, PLK2, ACP3, R3HDM2, NEAT1, CAMSAP1, DUSP2, DDIT3, ZFC3H1, RASGEF1B, SIK1, HBEGF, DUSP1, DUSP4, DAPK1, EGF, EGR1, ELF2, CTTN, UHRF1BP1L, ETV6, GADD45A, ZFAND3, APOBEC3A, UBE2E1, TNIK, SLC2A3, C9orf72, HLA-DRB1, PTPN22, IL6, IL2RA, STAT4, VTCN1, MIF, LACC1, PTPN2, IL23R, CD247, ACP5, ANKRD55, REEP3, ZMIZ1, IL2RB, TIMMDC1, ATG16L1, CRB1, ADGRL2, SUOX, ATXN2L, ERAP2, TNF, IGF2-AS, NRBF2, FNBP1, LINC00993, CARD9, CTTNBP2, CACNA1I, ANGPTL5, HLA-A, DAG1, IL1B, IL1A, FUT2, TSBP1, CCDC26, RBM45, GPR35, NKD1, LRRK2, HLA-DPB1, FAM169B, ANKRD30A, LURAP1L, C1orf141, DCLRE1C, CSMD1, TSBP1-AS1, LINC01250, INS-IGF2, IL10, IRF1-AS1, TNFSF15, CCR5, FOXP3, IFNG, IL4, IL17A, HLA-DQA1, TRAF1, CRP, ESR1, IL2, S100A12, CXCR3, NLRP3, DEK, MEFV, IL18, TLR4, ERAL1, IL6R, ISG20, TNFRSF11B, CRYGD, GZMB, UNC13D, MTHFR, GH1, CXCL8, HSPD1, MIR146A, VDR, MMP9, TRBV20OR9-2, COMP, CD226, TNFSF11, TNFRSF1A, CCR4, MMP3, NLRP1, TAP1, IL6ST, TLR2, TIMP2, TNFAIP3, LINC01193, IRF1, USO1, MBL2, SPP1, CXCL10, MIR155, TIMP1, IRF5, HLA-C, HLA-B, CSF2, IL33, PSMA6, PSMB9, NXF1, PRKCQ, VIL1, HLA-DQB1, VEGFA, IL21, S100A8, HLA-DRB3, HMGB1, HSPA14, SEC14L2, CCL3, CCL5, TNFRSF11A, IGF1, FLNB, MTX1, CXCR6, ABCB6, POSTN, NR1I3, SH2B3, HMGB1P5, TRIM13, NAMPT, CARD14, CCL27, SUMO4, NT5C1A, EBPL, FCRL3, HT, PYDC1, FAM177A1, SERPINA2, CARD8, MIR125A, MIR204, MIR21, WG, CXADRP1, KIR2DS2, TLR10, NAA25, CXCL16, ZNF395, RSBN1, SIAE, TREM1, ERAP1, IL23A, TLR7, LINC00328, MBL3P, CD274, IL37, ACAD8, PADI4, CLEC16A, BMS1, ACACA, HDAC9, GSTT1, HMOX1, HLA-G, HLA-DRB5, HLA-DPA1, HIF1A, HFE, GSTP1, HYAL1, GSTM1, GHR, GEM, GATA3, GAS6, FCN2, HNF4A, ICAM1, ELK3, ITPA, KIR2DS4, KIR2DS1, KIR2DL3, KIR2DL2, KIR2DL1, KIF5A, IRAK1, IFN1@, IL13, IL7R, IL4R, IGHG3, IFNA13, IFNA1, FBN1, SLC26A2, KIR3DL2, ARR3, C2, BMP4, BGLAP, BCL2, ATM, ATIC, AR, C4BPA, FAS, ANXA11, AMH, ACAN, ADRB2, ADA, C4A, C5, ATN1, CCR7, DPP4, DHCR7, CXADR, CTNNB1, CSF3, CNR2, CHIT1, CALCR, CDK6, LRBA, ENTPD1, CD28, CAT, CASR, KIR3DL1, KIR3DS1, PRORP, SNAI1, TGFB1, PRDX2, TAP2, STXBP2, STAT1, SPG7, SLC19A1, TLR3, SELP, SELE, CXCL11, CXCL6, CCL21, CCL20, TGM2, TNFRSF1B, ATXN2, ABCC3, NCR2, IL32, PSTPIP1, SH2D2A, KSR1, CCN6, DGKZ, TP53, CDR3, VIM, VCAM1, TYK2, TWIST1, TTN, CCL19, SAG, AFF3, MME, NGF, NFKBIA, NCAM1, MMP8, MMP2, MMP1, CIITA, NOS3, MECP2, LYZ, LTA, LIG4, LEP, LBP, NM, NT5E, SAA1, PTX3, S100B, S100A9, S100A1, BRD2, RBP3, RAB27A, PTPRC, PDCD1, PSMC6, PSMA3, PRNP, PRKAR1A, PRF1, ABCB1, CCL2
-
Pancreatic Neuroendocrine Tumor
Wikipedia
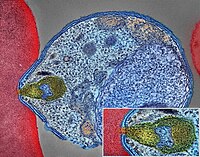
PanNETs are a type of neuroendocrine tumor , representing about one third of gastroenteropancreatic neuroendocrine tumors (GEP-NETs). Many PanNETs are benign , while some are malignant . ... Types of PNET based on hormones produced Type Relative incidence Typical location of tumor [5] Biomarkers [5] Symptoms [6] Insulinoma 35-40% [6] Head, body, tail of pancreas insulin, proinsulin , C-peptide Hypoglycemia Gastrinoma 16-30% [6] Gastrinoma triangle gastrin , PP Abdominal pain Refractory peptic ulcer disease Secretory diarrhea VIPoma <10% [6] Distal pancreas (body and tail) VIP Profuse watery diarrhea Dehydration Hypokalemia Achlorhydria Somatostatinoma <5% [6] Pancreatoduodenal groove, ampullary, periampullary somatostatin Diabetes mellitus Cholelithiasis Steatorrhea Anemia Weight loss PPoma Head or pancreas pancreatic polypeptide Glucagonoma 1% [7] Body and tail of pancreas glucagon , glycentin Necrolytic migratory erythema Diabetes mellitus Diarrhea Deep vein thrombosis Relative incidence is given as percentage of all functional pancreatic neuroendocrine tumors. ... Up to 60% [ medical citation needed ] of PanNETs are nonsecretory or nonfunctional, in which there is no secretion, or the quantity or type of products, such as pancreatic polypeptide (PPoma), chromogranin A, and neurotensin , do not cause a clinical syndrome although blood levels may be elevated. [10] In total, 85% of PanNETs have an elevated blood marker. [2] Functional tumors are often classified by the hormone most strongly secreted, for example: gastrinoma : the excessive gastrin causes Zollinger–Ellison syndrome (ZES) with peptic ulcers and diarrhea insulinoma : [11] hypoglycemia occurs with concurrent elevations of insulin , proinsulin and C peptide [12] glucagonoma : the symptoms are not all due to glucagon elevations, [12] and include a rash , sore mouth, altered bowel habits, venous thrombosis , and high blood glucose levels [12] VIPoma , producing excessive vasoactive intestinal peptide , which may cause profound chronic w atery d iarrhea and resultant dehydration , h ypokalemia , and a chlorhydria (WDHA or pancreatic cholera syndrome) somatostatinoma : these rare tumors are associated with elevated blood glucose levels, achlorhydria , cholelithiasis , and diarrhea [12] less common types include ACTHoma , CRHoma , calcitoninoma , GHRHoma , GRFoma , and parathyroid hormone–related peptide tumor In these various types of functional tumors, the frequency of malignancy and the survival prognosis have been estimated dissimilarly, but a pertinent accessible summary is available. [13] Diagnosis [ edit ] Because symptoms are non-specific, diagnosis is often delayed. [14] Measurement of hormones including pancreatic polypeptide , gastrin , proinsulin , insulin , glucagon , and vasoactive intestinal peptide can determine if a tumor is causing hypersecretion. [14] [15] Multiphase CT and MRI are the primary modalities for morphologic imaging of PNETs. ... However, morphological imaging alone is not sufficient for a definite diagnosis [14] [16] On biopsy , immunohistochemistry is generally positive for chromogranin and synaptophysin . [17] Genetic testing thereof typically shows altered MEN1 and DAXX / ATRX . [17] Staging [ edit ] The 2010 WHO classification of tumors of the digestive system grades all the neuroendocrine tumors into three categories, based on their degree of cellular differentiation (from well-differentiated "NET G1" through to poorly-differentiated "NET G3"). ... Combinations of several medicines have been used, such as doxorubicin with streptozocin and fluorouracil (5-FU) [12] and capecitabine with temozolomide. [ citation needed ] Although marginally effective in well-differentiated PETs, cisplatin with etoposide has some activity in poorly differentiated neuroendocrine cancers (PDNECs), [12] particularly if the PDNEC has an extremely high Ki-67 score of over 50%. [8] : 30 Several targeted therapy agents have been approved in PanNETs by the FDA based on improved progression-free survival (PFS): everolimus (Afinitor) is labeled for treatment of progressive neuroendocrine tumors of pancreatic origin in patients with unresectable, locally advanced or metastatic disease. [20] [21] The safety and effectiveness of everolimus in carcinoid tumors have not been established. [20] [21] sunitinib (Sutent) is labeled for treatment of progressive, well-differentiated pancreatic neuroendocrine tumors in patients with unresectable locally advanced or metastatic disease. [22] [23] Sutent also has approval from the European Commission for the treatment of 'unresectable or metastatic, well-differentiated pancreatic neuroendocrine tumors with disease progression in adults'. [24] A phase III study of sunitinib treatment in well differentiated pNET that had worsened within the past 12 months (either advanced or metastatic disease) showed that sunitinib treatment improved progression-free survival (11.4 months vs. 5.5 months), overall survival , and the objective response rate (9.3% vs. 0.0%) when compared with placebo. [25] Genetics [ edit ] Pancreatic neuroendocrine tumors may arise in the context of multiple endocrine neoplasia type 1 , Von Hippel–Lindau disease , neurofibromatosis type 1 (NF-1) or tuberose sclerosis (TSC) [26] [27] Analysis of somatic DNA mutations in well-differentiated pancreatic neuroendocrine tumors identified four important findings: [28] [6] as expected, the genes mutated in NETs, MEN1 , ATRX , DAXX , TSC2 , PTEN and PIK3CA , [28] are different from the mutated genes previously found in pancreatic adenocarcinoma . [29] [30] one in six well-differentiated pancreatic NETs have mutations in mTOR pathway genes, such as TSC2 , PTEN and PIK3CA . [28] The sequencing discovery might allow selection of which NETs would benefit from mTOR inhibition such as with everolimus , but this awaits validation in a clinical trial . mutations affecting a new cancer pathway involving ATRX and DAXX genes were found in about 40% of pancreatic NETs. [28] The proteins encoded by ATRX and DAXX participate in chromatin remodeling of telomeres ; [31] these mutations are associated with a telomerase -independent maintenance mechanism termed ALT (alternative lengthening of telomeres) that results in abnormally long telomeric ends of chromosomes . [31] ATRX / DAXX and MEN1 mutations were associated with a better prognosis . [28] References [ edit ] ^ Burns WR, Edil BH (March 2012).
-
Retiform Parapsoriasis
Wikipedia
Retiform parapsoriasis Specialty Dermatology Retiform parapsoriasis is a cutaneous condition, considered to be a type of large-plaque parapsoriasis . [1] It is characterized by widespread, ill-defined plaques on the skin, that have a net-like or zebra-striped pattern. [2] Skin atrophy , a wasting away of the cutaneous tissue , usually occurs within the area of these plaques. [1] See also [ edit ] Parapsoriasis Poikiloderma vasculare atrophicans List of cutaneous conditions References [ edit ] ^ a b Lambert WC, Everett MA (Oct 1981). ... External links [ edit ] Classification D ICD - 10 : L41.5 ICD - 9-CM : 696.2 v t e Papulosquamous disorders Psoriasis Pustular Generalized pustular psoriasis ( Impetigo herpetiformis ) Acropustulosis / Pustulosis palmaris et plantaris ( Pustular bacterid ) Annular pustular psoriasis Localized pustular psoriasis Other Guttate psoriasis Psoriatic arthritis Psoriatic erythroderma Drug-induced psoriasis Inverse psoriasis Napkin psoriasis Seborrheic-like psoriasis Parapsoriasis Pityriasis lichenoides ( Pityriasis lichenoides et varioliformis acuta , Pityriasis lichenoides chronica ) Lymphomatoid papulosis Small plaque parapsoriasis ( Digitate dermatosis , Xanthoerythrodermia perstans ) Large plaque parapsoriasis ( Retiform parapsoriasis ) Other pityriasis Pityriasis rosea Pityriasis rubra pilaris Pityriasis rotunda Pityriasis amiantacea Other lichenoid Lichen planus configuration Annular Linear morphology Hypertrophic Atrophic Bullous Ulcerative Actinic Pigmented site Mucosal Nails Peno-ginival Vulvovaginal overlap synromes with lichen sclerosus with lupus erythematosis other: Hepatitis-associated lichen planus Lichen planus pemphigoides Other Lichen nitidus Lichen striatus Lichen ruber moniliformis Gianotti–Crosti syndrome Erythema dyschromicum perstans Idiopathic eruptive macular pigmentation Keratosis lichenoides chronica Kraurosis vulvae Lichen sclerosus Lichenoid dermatitis Lichenoid reaction of graft-versus-host disease This dermatology article is a stub .
-
Skin/hair/eye Pigmentation, Variation In, 9
OMIM
The binding of ASP to MSHR precludes alpha-MSH-initiated signaling and thus blocks production of cAMP, leading to a downregulation of eumelanogenesis. The net result is increased synthesis of pheomelanin. ... A total of 6 SNPs within a region of strong linkage disequilibrium on 20q11.22 showed association with burning and freckling that reached genomewide significance (max odds ratio = 1.60, p = 3.9 x 10(-9)). Multipoint analysis of the area revealed an extended haplotype tagged by a 2-SNP haplotype, rs1015362G and rs4911414T, that Sulem et al. (2008) referred to as 'the ASIP haplotype.' ... The strength of the association of rs6058017 with the pigmentation traits was much less than that of the ASIP haplotype, and after adjustment for rs6058017 the ASIP haplotype remained highly significant for burning and freckling (p = 1.3 x 10(-46)). After adjustment for the ASIP haplotype, the association of rs6058017 with pigmentation characteristics was only marginal (P = 0.057 for burning and freckling).
-
Leishmaniasis
Wikipedia
Servicio Jesuita a Refugiados. Archived from the original on 10 November 2005. ^ "CENTRAL/S. ASIA – Kabul: A city in intensive care" . ... Reuters. Archived from the original on 10 December 2015 . Retrieved 8 December 2015 . ^ Birsel R (28 June 2002). ... Archived from the original on 13 February 2011 . Retrieved 10 February 2011 . ^ "Business: Company's mesh will help troops beat 'Baghdad boils ' " . ... Archived from the original on 6 June 2010 . Retrieved 10 February 2011 . ^ Srivastava S, Shankar P, Mishra J, Singh S (May 2016). ... International Journal of Preventive Medicine . 10 : 95. doi : 10.4103/ijpvm.IJPVM_116_18 .TNF, IFNG, IL10, IL6, ARG1, IL18, CRP, TNFRSF18, MCL1, HSPA4, IL1B, SLC11A1, CXCL10, NLRP3, IL17A, TLR2, CCR5, TLR4, IL32, PRDX2, LEP, TGFB1, CD274, FCN2, CD163, MTOR, HM13, IL4, BCL2, BAX, LMLN, IGF1, HIF1A, ANXA1, VDR, UNG, TAM, NR0B2, EZR, ADA, TLR3, STAT1, MAPK3, MAPK4, EIF2AK2, PSG5, PSMD7, PTHLH, PTPN1, PTPN2, PTPN6, RPA1, RPS6, CCL2, CCL8, CXCL11, SLC1A5, SLC1A7, SNAP25, SOAT1, SPP1, TP63, EIF2S2, CDK5R1, GOPC, FOXP3, HSPA14, CD244, TOLLIP, FBLIM1, MSTO1, FBXW7, ACSS2, PDXP, SLC52A2, ALDH1A2, TMPRSS13, DCLK3, IL33, CDCA5, PWAR1, ARMH1, HNP1, CCR2, UPK3B, DLL1, SGSM3, NOX1, PABPC1, NR1I2, SPHK1, EIF2B4, EIF2B2, PRKAB1, HSPB3, SLC7A6, ARHGEF2, AIM2, H6PD, RABEPK, LANCL1, TNFSF13B, EBNA1BP2, CD160, GABARAPL2, GABARAPL1, PRDX5, POLR1A, MAPK1, NOS2, PRKAA2, PRKAA1, CST3, CTLA4, CTSB, CTSL, CYP51A1, DDT, DHFR, DPAGT1, DPP4, DSPP, DUSP4, EEF1B2, EEF2, EGFR, EIF2B1, F2R, FCGR2A, FECH, FLI1, CPB1, CCR7, LRBA, ATR, AKT1, ALDH1A1, APEX1, APRT, AQP1, ATM, ATP2A3, ATP2B4, PRDM1, CD69, BRCA1, CAPN1, CD1A, CD28, CD86, CD40, CD40LG, CD44, FPR2, G6PD, GAPDH, CYTB, MNAT1, CD200, MPG, MPL, MPST, MRC1, MSMB, MST1, AHR, MFAP1, PAEP, PHB, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PLP1, PNOC, MAP3K10, MBL2, GCHFR, IFNB1, GCK, GTF3C1, HLA-C, HMOX1, HSPD1, IFN1@, IFNA1, IFNA13, IL1A, LTA, IL9, IL12A, IL12RB1, IL13, ITGA4, ITGAL, JAK2, RPSA, H3P28
-
Lymphatic Filariasis
Wikipedia
In areas endemic for podoconiosis, prevalence can be 5% or higher. [34] In communities where lymphatic filariasis is endemic, as many as 10% of women can be afflicted with swollen limbs, and 50% of men can suffer from mutilating genital symptoms. [20] Filariasis is considered endemic in 73 countries; 37 of these are in Africa. ... Archived from the original on 2016-10-12. ^ a b c d e f g h i j k l m n o p q r s t u "Lymphatic filariasis Fact sheet N°102" . ... PLOS Neglected Tropical Diseases . 2 (10): e317. doi : 10.1371/journal.pntd.0000317 . ... PLOS Neglected Tropical Diseases . 7 (10): e2301. doi : 10.1371/journal.pntd.0002301 . ... Archived from the original on 2008-12-10 . Retrieved 2008-11-21 . ^ Grove, David I (1990).
-
West Nile Fever
Wikipedia
Centers for Disease Control and Prevention . USA.gov. 2018-12-10 . Retrieved 15 January 2019 . ^ "West Nile virus" . ... "West Nile virus infection and conjunctive exposure" . Emerging Infect. Dis . 11 (10): 1648–9. doi : 10.3201/eid1110.040212 . ... Retrieved 24 August 2016 . ^ "Prevention | West Nile Virus | CDC" . www.cdc.gov . 2018-09-24 . Retrieved 2018-10-29 . ^ a b Rios L, Maruniak JE (October 2011). ... Journal of Virological Methods . 51 (2–3): 201–10. doi : 10.1016/0166-0934(94)00105-P . ... "Long-term prognosis for clinical West Nile virus infection" . Emerg Infect Dis . 10 (8): 1405–11. doi : 10.3201/eid1008.030879 .CCR5, ERVK-32, ROBO3, MAVS, DDX58, PLAAT4, IFIT2, ERVK-6, STAT1, SPP1, OAS1, IL1B, IFNB1, RNASEL, CASP8, HLA-DRB1, PELI1, SELENBP1, ARHGEF2, LRRFIP1, NAMPT, TRAIP, RIPK3, SEC14L2, CSF1R, LAMP3, ERVW-1, FOXP3, ZMYND10, DDX56, CCR7, VCP, CDKN2A, IFIH1, DHX58, ZBP1, HAVCR2, PIK3IP1, NLRP3, TNFRSF13C, TRIM6, RBM45, CCR2, ERVK-20, ERVK-18, VAMP8, TNFRSF1A, IFNA1, TNF, IFNA13, HLA-DQA1, IL1A, HLA-C, IL10, IL17A, IL18, IRF3, IRF5, KIR2DL2, KIR3DL1, KIR3DS1, LSAMP, CD180, SMAD4, MMP9, HLA-A, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PZP, GLS, CASP1, SNCA, GEM, DDX3X, TAP1, TLR3, ATF4
-
Visceral Leishmaniasis
Wikipedia
Depletion of CD8+ T cells from VL PBMC stopped endogenous IL-10 secretion but increased Leishmania antigen specific IL-10 secretion, suggesting that CD8+ regulatory T cells are responsible for endogenous IL-10 secretion. [27] CD4+ clones could only be isolated from VL PBMC after CD8+ T cell depletion. ... If possible, use a bed net that has been soaked in or sprayed with a pyrethroid-containing insecticide. ... PLOS Neglected Tropical Diseases . 10 (11): e0005150. doi : 10.1371/journal.pntd.0005150 . ... "Role of CD8+ T Cells in Endogenous Interleukin-10 Secretion Associated with Visceral Leishmaniasis" . ... J Immunol . 196 (10): 4100–9. doi : 10.4049/jimmunol.1502678 .TNF, IL10, IFNG, SLC11A1, IL1B, HLA-DRB1, IL6, HMOX1, CSF2, HLA-DQA1, CXCL8, IL10RA, CRP, IL4, HSPA4, TLR4, MBL2, FOXP3, HIF1A, SLC6A3, TLR2, LEP, IL17A, IL9, IL2, TLR9, P4HB, CYTB, IL15, LMLN, IL12RB1, DDX56, IGF1, CCR7, CD69, MAPK1, ODC1, DLL1, RAB6A, MPL, NOS2, IL4R, CD58, IFNGR1, RCE1, NUP93, NPEPPS, PDCD1LG2, RABEPK, IL32, UNC13D, CPQ, ZNRD2, CXCL13, DCTN6, TICAM2, LANCL1, SOCS1, ARHGEF2, TGFB1, SMPD2, SPAG1, ST2, STAT3, TARS1, TBP, TGFBI, PKD2L1, H3P23, TP53, TNFSF4, TP63, POP1, HSPB3, TNFSF13B, SMUG1, EBNA1BP2, MON2, MIR155, CDK12, TREM1, ZNF469, FBLIM1, GOPC, HAMP, MIR122, CXCL16, AGMO, ASPG, NOD2, FAM120B, POPDC3, MLST8, PDIA2, TARS2, TMED7-TICAM2, IL33, IL23A, CLEC4D, DICER1, SIRT1, PIGZ, SLC17A5, CD274, RBM45, PADI1, PYDC5, NELFCD, PSAT1, TARS3, NLRP3, SMARCB1, MTCO2P12, TMED7, PRRT2, ARMH1, ACTB, SLAMF1, SGTA, F5, FCN2, FH, FOXO1, G6PD, GABPA, HLA-C, HLA-G, HSPA1A, HSPA1B, HSPA2, HSP90AA1, IFI27, IFNA1, IFNA13, IL2RA, IL2RB, ELF2, EEF1B2, DUSP4, CD14, AHCY, APOA1, KLK3, AQP1, BCS1L, CASP3, CD1A, CD34, DPP4, CD40, CD40LG, CPB1, CTLA4, CTSB, CYP51A1, DDT, IL5, CXCR1, CXCR2, PPARA, NCAM1, NFE2L2, NOTCH3, NRAS, ADA, PHB, PLAG1, PRKCA, COX2, PRKCB, MAPK10, PROS1, PSMD7, PSMD9, RNASE2, CCL2, MYD88, MMP9, IL13, RPSA, IL18, CXCL10, INPP5D, IRF1, IRF5, ISG20, ITGB2, LCK, MMP2, LECT2, LGALS1, LPL, LTA, MATK, MIF, CXCL9, H3P28
-
Nephrolithiasis, Calcium Oxalate
OMIM
In populations of European ancestry, 5 to 10% of adults experience the painful precipitation of calcium oxalate in their urinary tracts. ... Several presumed carrier females did not have calculi. Fifteen males (and no females) in 10 sibships were affected. The systematic genetic study of calcium oxalate renal calculi done by Resnick et al. (1968) led to the conclusion that monogenic inheritance could be excluded; the findings were considered compatible with the hypothesis that the tendency to form calcium oxalate renal stones is regulated by a polygenic system, with less risk for females than males. ... In vitro flux studies indicated that mice lacking Slc26a6 have a defect in intestinal oxalate secretion resulting in enhanced net absorption of oxalate. Jiang et al. (2006) concluded that the anion exchanger, SLC26A6, has a major constitutive role in limiting net intestinal absorption of oxalate, thereby preventing hyperoxaluria and calcium oxalate urolithiasis.