-
Noonan Syndrome
Wikipedia
Genetic condition involving facial, heart, blood and skeletal features Noonan syndrome Other names Male Turner syndrome, Noonan-Ehmke syndrome, Turner-like syndrome, Ullrich-Noonan syndrome [1] A 12-year-old girl with Noonan syndrome. ... In Turner syndrome, there is a lower incidence of developmental delays, left-sided heart defects are constant and the occurrence of renal abnormalities is much lower. [20] Other RASopathies Watson syndrome - Watson Syndrome has a number of similar characteristics with Noonan's Syndrome such as short stature, pulmonary valve stenosis, variable intellectual development, and skin pigment changes. [20] [21] Cardiofaciocutaneous (CFC) syndrome - CFC syndrome is very similar to Noonan's Syndrome due to similar cardiac and lymphatic features. However, In CFC syndrome intellectual disability and gastrointestinal problems are often more severe and pronounced. [20] [22] Costello syndrome - Like CFC syndrome, Costello syndrome has overlapping features with Noonan's Syndrome. ... References [ edit ] ^ a b c d e f g h i j k l m "Noonan syndrome" . Genetics Home Reference . Retrieved 24 December 2018 . ^ a b c d e f g "Noonan Syndrome" . ... External links [ edit ] Classification D ICD - 10 : Q87.1 ICD - 9-CM : 759.89 OMIM : 163950 605275 609942 610733 611553 MeSH : D009634 DiseasesDB : 29094 SNOMED CT : 205824006 External resources MedlinePlus : 001656 eMedicine : article/947504 Patient UK : Noonan syndrome GeneReviews : Noonan Syndrome Orphanet : 648 Wikimedia Commons has media related to Noonan syndrome . v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome v t e Deficiencies of intracellular signaling peptides and proteins GTP-binding protein regulators GTPase-activating protein Neurofibromatosis type I Watson syndrome Tuberous sclerosis Guanine nucleotide exchange factor Marinesco–Sjögren syndrome Aarskog–Scott syndrome Juvenile primary lateral sclerosis X-Linked mental retardation 1 G protein Heterotrimeic cAMP / GNAS1 : Pseudopseudohypoparathyroidism Progressive osseous heteroplasia Pseudohypoparathyroidism Albright's hereditary osteodystrophy McCune–Albright syndrome CGL 2 Monomeric RAS: HRAS Costello syndrome KRAS Noonan syndrome 3 KRAS Cardiofaciocutaneous syndrome RAB: RAB7 Charcot–Marie–Tooth disease RAB23 Carpenter syndrome RAB27 Griscelli syndrome type 2 RHO: RAC2 Neutrophil immunodeficiency syndrome ARF : SAR1B Chylomicron retention disease ARL13B Joubert syndrome 8 ARL6 Bardet–Biedl syndrome 3 MAP kinase Cardiofaciocutaneous syndrome Other kinase / phosphatase Tyrosine kinase BTK X-linked agammaglobulinemia ZAP70 ZAP70 deficiency Serine/threonine kinase RPS6KA3 Coffin-Lowry syndrome CHEK2 Li-Fraumeni syndrome 2 IKBKG Incontinentia pigmenti STK11 Peutz–Jeghers syndrome DMPK Myotonic dystrophy 1 ATR Seckel syndrome 1 GRK1 Oguchi disease 2 WNK4 / WNK1 Pseudohypoaldosteronism 2 Tyrosine phosphatase PTEN Bannayan–Riley–Ruvalcaba syndrome Lhermitte–Duclos disease Cowden syndrome Proteus-like syndrome MTM1 X-linked myotubular myopathy PTPN11 Noonan syndrome 1 LEOPARD syndrome Metachondromatosis Signal transducing adaptor proteins EDARADD EDARADD Hypohidrotic ectodermal dysplasia SH3BP2 Cherubism LDB3 Zaspopathy Other NF2 Neurofibromatosis type II NOTCH3 CADASIL PRKAR1A Carney complex PRKAG2 Wolff–Parkinson–White syndrome PRKCSH PRKCSH Polycystic liver disease XIAP XIAP2 See also intracellular signaling peptides and proteins Authority control GND : 4171991-8PTPN11, RAF1, KRAS, SOS1, LZTR1, RIT1, SHOC2, NRAS, BRAF, MAP2K1, HRAS, MAP2K2, SOS2, RRAS2, A2ML1, MRAS, RRAS, RASA2, CBL, NF1, PPP1CB, KAT6B, EPHA2, RASA1, CENPJ, ATR, CEP63, APAF1, EPHB2, GH1, MAPK1, ZHX2, PTEN, ACP1, PTPRU, REG1A, SLC25A3, IGF1, MAPK3, CDC42, STAT5B, XYLT2, MUL1, MVD, MAP2K7, CBLL2, PRKN, CDAN1, MED18, FHL1, ELK1, DTX1, MIR195, SGSM3, SPRED1, CDC25C, AMH, SLC2A4RG, ZNF462, PTPRT, DSG4, ARAF, MTG1, MAPK7, GRIN1, ARHGEF2, MPZL1, PTPN1, PDGFRB, NOTCH1, ACVR1, RASGRF1, MYC, RNASE3, RPL6, RPS6KA3, CCL3, SHOX, IGFBP3, STAT3, STAT5A, USF1, GRIN2B, NCK2, MIR223
-
Charcot–marie–tooth Disease
Wikipedia
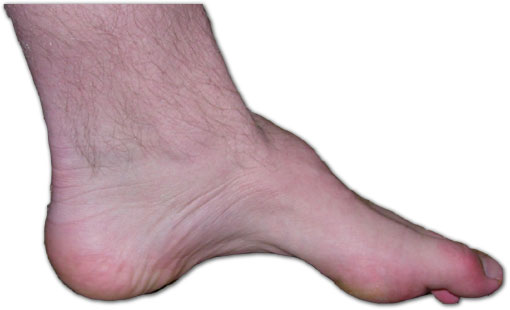
Please improve this by adding secondary or tertiary sources . ( May 2020 ) ( Learn how and when to remove this template message ) Charcot-Marie-Tooth disease Other names Charcot–Marie–Tooth neuropathy, peroneal muscular atrophy, Dejerine-Sottas Syndrome The foot of a person with Charcot–Marie–Tooth disease: The lack of muscle, a high arch , and claw toes are signs of this genetic disease. ... When neuropathic pain is present as a symptom of CMT, it is comparable to that seen in other peripheral neuropathies , as well as postherpetic neuralgia and complex regional pain syndrome , among other diseases. [9] Causes [ edit ] Chromosome 17 Charcot–Marie–Tooth disease is caused by genetic mutations that cause defects in neuronal proteins. ... "Gastrointestinal involvement in neurologic disorders: Stiff-man and Charcot–Marie–Tooth syndromes". The American Journal of the Medical Sciences . 313 (1): 70–73. doi : 10.1097/00000441-199701000-00012 . ... Retrieved August 26, 2011 . ^ "Charcot-Marie-Tooth Syndrome. CMT information" . Patient . ^ Carter, Gregory T.; Jensen, Mark P.; Galer, Bradley S.; Kraft, George H.; Crabtree, Linda D.; Beardsley, Ruth M.; Abresch, Richard T.; Bird, Thomas D. (1998). ... External links [ edit ] Classification D ICD - 10 : G60.0 ICD - 9-CM : 356.1 OMIM : 311860 611566 311070 605588 311850 (X Type 5) 302800 304040 (X Type 1) MeSH : D002607 DiseasesDB : 5815 External resources MedlinePlus : 000727 eMedicine : orthoped/43 pmr/29 GeneReviews : Charcot-Marie-Tooth Hereditary Neuropathy Overview Charcot-Marie-Tooth Neuropathy Type 1 Charcot-Marie-Tooth Neuropathy X Type 5 Charcot-Marie-Tooth Neuropathy X Type 1 GARS-Associated Axonal Neuropathy, Charcot-Marie-Tooth Neuropathy Type 2D, Distal Spinal Muscular Atrophy V Charcot–Marie–Tooth disease at Curlie v t e Diseases relating to the peripheral nervous system Mononeuropathy Arm median nerve Carpal tunnel syndrome Ape hand deformity ulnar nerve Ulnar nerve entrapment Froment's sign Ulnar tunnel syndrome Ulnar claw radial nerve Radial neuropathy Wrist drop Cheiralgia paresthetica long thoracic nerve Winged scapula Backpack palsy Leg lateral cutaneous nerve of thigh Meralgia paraesthetica tibial nerve Tarsal tunnel syndrome plantar nerve Morton's neuroma superior gluteal nerve Trendelenburg's sign sciatic nerve Piriformis syndrome Cranial nerves See Template:Cranial nerve disease Polyneuropathy and Polyradiculoneuropathy HMSN Charcot–Marie–Tooth disease Dejerine–Sottas disease Refsum's disease Hereditary spastic paraplegia Hereditary neuropathy with liability to pressure palsy Familial amyloid neuropathy Autoimmune and demyelinating disease Guillain–Barré syndrome Chronic inflammatory demyelinating polyneuropathy Radiculopathy and plexopathy Brachial plexus injury Thoracic outlet syndrome Phantom limb Other Alcoholic polyneuropathy Other General Complex regional pain syndrome Mononeuritis multiplex Peripheral neuropathy Neuralgia Nerve compression syndrome v t e Inherited disorders of trafficking / vesicular transport proteins Vesicle formation Lysosome / Melanosome : HPS1 – HPS7 Hermansky–Pudlak syndrome LYST Chédiak–Higashi syndrome COPII : SEC23A Cranio-lenticulo-sutural dysplasia COG7 CDOG IIE APC: AP1S2 X-linked intellectual disability AP3B1 Hermansky–Pudlak syndrome 2 AP4M1 CPSQ3 Rab ARL6 BBS3 RAB27A Griscelli syndrome 2 CHM Choroideremia MLPH Griscelli syndrome 3 Cytoskeleton Myosin : MYO5A Griscelli syndrome 1 Microtubule : SPG4 Hereditary spastic paraplegia 4 Kinesin : KIF5A Hereditary spastic paraplegia 10 Spectrin : SPTBN2 Spinocerebellar ataxia 5 Vesicle fusion Synaptic vesicle : SNAP29 CEDNIK syndrome STX11 Hemophagocytic lymphohistiocytosis 4 Caveolae : CAV1 Congenital generalized lipodystrophy 3 CAV3 Limb-girdle muscular dystrophy 2B , Long QT syndrome 9 Vacuolar protein sorting : VPS33B ARC syndrome VPS13B Cohen syndrome DYSF Distal muscular dystrophy See also vesicular transport proteins v t e Cytoskeletal defects Microfilaments Myofilament Actin Hypertrophic cardiomyopathy 11 Dilated cardiomyopathy 1AA DFNA20 Nemaline myopathy 3 Myosin Elejalde syndrome Hypertrophic cardiomyopathy 1, 8, 10 Usher syndrome 1B Freeman–Sheldon syndrome DFN A3, 4, 11, 17, 22; B2, 30, 37, 48 May–Hegglin anomaly Troponin Hypertrophic cardiomyopathy 7, 2 Nemaline myopathy 4, 5 Tropomyosin Hypertrophic cardiomyopathy 3 Nemaline myopathy 1 Titin Hypertrophic cardiomyopathy 9 Other Fibrillin Marfan syndrome Weill–Marchesani syndrome Filamin FG syndrome 2 Boomerang dysplasia Larsen syndrome Terminal osseous dysplasia with pigmentary defects IF 1/2 Keratinopathy ( keratosis , keratoderma , hyperkeratosis ): KRT1 Striate palmoplantar keratoderma 3 Epidermolytic hyperkeratosis IHCM KRT2E ( Ichthyosis bullosa of Siemens ) KRT3 ( Meesmann juvenile epithelial corneal dystrophy ) KRT4 ( White sponge nevus ) KRT5 ( Epidermolysis bullosa simplex ) KRT8 ( Familial cirrhosis ) KRT10 ( Epidermolytic hyperkeratosis ) KRT12 ( Meesmann juvenile epithelial corneal dystrophy ) KRT13 ( White sponge nevus ) KRT14 ( Epidermolysis bullosa simplex ) KRT17 ( Steatocystoma multiplex ) KRT18 ( Familial cirrhosis ) KRT81 / KRT83 / KRT86 ( Monilethrix ) Naegeli–Franceschetti–Jadassohn syndrome Reticular pigmented anomaly of the flexures 3 Desmin : Desmin-related myofibrillar myopathy Dilated cardiomyopathy 1I GFAP : Alexander disease Peripherin : Amyotrophic lateral sclerosis 4 Neurofilament : Parkinson's disease Charcot–Marie–Tooth disease 1F, 2E Amyotrophic lateral sclerosis 5 Laminopathy : LMNA Mandibuloacral dysplasia Dunnigan Familial partial lipodystrophy Emery–Dreifuss muscular dystrophy 2 Limb-girdle muscular dystrophy 1B Charcot–Marie–Tooth disease 2B1 LMNB Barraquer–Simons syndrome LEMD3 Buschke–Ollendorff syndrome Osteopoikilosis LBR Pelger–Huet anomaly Hydrops-ectopic calcification-moth-eaten skeletal dysplasia Microtubules Kinesin Charcot–Marie–Tooth disease 2A Hereditary spastic paraplegia 10 Dynein Primary ciliary dyskinesia Short rib-polydactyly syndrome 3 Asphyxiating thoracic dysplasia 3 Other Tauopathy Cavernous venous malformation Membrane Spectrin : Spinocerebellar ataxia 5 Hereditary spherocytosis 2, 3 Hereditary elliptocytosis 2, 3 Ankyrin : Long QT syndrome 4 Hereditary spherocytosis 1 Catenin APC Gardner's syndrome Familial adenomatous polyposis plakoglobin ( Naxos syndrome ) GAN ( Giant axonal neuropathy ) Other desmoplakin : Striate palmoplantar keratoderma 2 Carvajal syndrome Arrhythmogenic right ventricular dysplasia 8 plectin : Epidermolysis bullosa simplex with muscular dystrophy Epidermolysis bullosa simplex of Ogna plakophilin : Skin fragility syndrome Arrhythmogenic right ventricular dysplasia 9 centrosome : PCNT ( Microcephalic osteodysplastic primordial dwarfism type II ) Related topics: Cytoskeletal proteins v t e Cell membrane protein disorders (other than Cell surface receptor , enzymes , and cytoskeleton ) Arrestin Oguchi disease 1 Myelin Pelizaeus–Merzbacher disease Dejerine–Sottas disease Charcot–Marie–Tooth disease 1B, 2J Pulmonary surfactant Surfactant metabolism dysfunction 1, 2 Cell adhesion molecule IgSF CAM : OFC7 Cadherin : DSG1 Striate palmoplantar keratoderma 1 DSG2 Arrhythmogenic right ventricular dysplasia 10 DSG4 LAH1 DSC2 Arrhythmogenic right ventricular dysplasia 11 Integrin : cell surface receptor deficiencies Tetraspanin TSPAN7 X-Linked mental retardation 58 TSPAN12 Familial exudative vitreoretinopathy 5 Other KIND1 Kindler syndrome HFE HFE hereditary haemochromatosis DYSF Distal muscular dystrophy Limb-girdle muscular dystrophy 2B See also other cell membrane proteins v t e Deficiencies of intracellular signaling peptides and proteins GTP-binding protein regulators GTPase-activating protein Neurofibromatosis type I Watson syndrome Tuberous sclerosis Guanine nucleotide exchange factor Marinesco–Sjögren syndrome Aarskog–Scott syndrome Juvenile primary lateral sclerosis X-Linked mental retardation 1 G protein Heterotrimeic cAMP / GNAS1 : Pseudopseudohypoparathyroidism Progressive osseous heteroplasia Pseudohypoparathyroidism Albright's hereditary osteodystrophy McCune–Albright syndrome CGL 2 Monomeric RAS: HRAS Costello syndrome KRAS Noonan syndrome 3 KRAS Cardiofaciocutaneous syndrome RAB: RAB7 Charcot–Marie–Tooth disease RAB23 Carpenter syndrome RAB27 Griscelli syndrome type 2 RHO: RAC2 Neutrophil immunodeficiency syndrome ARF : SAR1B Chylomicron retention disease ARL13B Joubert syndrome 8 ARL6 Bardet–Biedl syndrome 3 MAP kinase Cardiofaciocutaneous syndrome Other kinase / phosphatase Tyrosine kinase BTK X-linked agammaglobulinemia ZAP70 ZAP70 deficiency Serine/threonine kinase RPS6KA3 Coffin-Lowry syndrome CHEK2 Li-Fraumeni syndrome 2 IKBKG Incontinentia pigmenti STK11 Peutz–Jeghers syndrome DMPK Myotonic dystrophy 1 ATR Seckel syndrome 1 GRK1 Oguchi disease 2 WNK4 / WNK1 Pseudohypoaldosteronism 2 Tyrosine phosphatase PTEN Bannayan–Riley–Ruvalcaba syndrome Lhermitte–Duclos disease Cowden syndrome Proteus-like syndrome MTM1 X-linked myotubular myopathy PTPN11 Noonan syndrome 1 LEOPARD syndrome Metachondromatosis Signal transducing adaptor proteins EDARADD EDARADD Hypohidrotic ectodermal dysplasia SH3BP2 Cherubism LDB3 Zaspopathy Other NF2 Neurofibromatosis type II NOTCH3 CADASIL PRKAR1A Carney complex PRKAG2 Wolff–Parkinson–White syndrome PRKCSH PRKCSH Polycystic liver disease XIAP XIAP2 See also intracellular signaling peptides and proteinsPMP22, GDAP1, MPZ, LRSAM1, FIG4, TRPV4, KIF1B, LMNA, GJB1, MFN2, SH3TC2, DYNC1H1, MORC2, HOXD10, SPTLC1, SBF2, SPTLC2, SETX, EGR2, MTMR2, SLC25A46, NEFL, GARS1, NDRG1, FGD4, DHTKD1, COX6A1, GSTT2, DNM2, SLC12A6, HSPB8, HSPB1, AARS1, ATP6, SBF1, MED25, ARHGEF10, IGHMBP2, MARS1, HINT1, DNAJB2, BAG3, PRX, MAD2L1BP, INF2, WASHC5, CTDP1, LITAF, RAB7B, RNMT, DES, RAB7A, TTN, GART, HSPB3, MTM1, YARS1, HSPB2, JPH1, YARS2, HDAC6, BSCL2, NEFH, PRPS1, HARS1, AIFM1, CMTX3, SMN1, FXN, PMP2, PLEK, PLEKHG5, PDK3, PRKN, CBLL2, MUL1, GNB4, ATP1A1, CANX, CCL2, SPG11, SMN2, DCPS, SOD1, TNF, MCM3AP, AARS2, TSG101, PNKP, SLURP1, RIEG2, SLA2, TFG, TUBA1B, SOX10, MME, CDC42, MAG, POLG, EGFR, KIF5A, APOA2, NEDD4, CNTF, HADHB, ACE, MFN1, DFNA7, DNM1, CUX1, SLC2A4RG, CSF1, DMPK, ROM1, DRP2, MYO15A, EEF1A1, IL17B, SIGLEC7, EMP2, EMP3, SACS, HARS2, TARDBP, TRIM2, IQSEC2, LPAR1, TEKT3, COX10, ERBB3, GSTT2B, GUSBP3, MIR29A, PLEKHM3, GSTK1, ACTG1, DFNA49, APCDD1, SLCO6A1, C1orf194, APP, LRIG3, AHNAK2, ARSA, SERPINC1, DGAT2, SYVN1, ATP7A, BAG1, SCARB2, CD44, CMTX2, ACD, ERBB2, PSIP1, RAB18, BUD31, NTF3, KHSRP, PFN2, GAN, MBS2, PGK1, D1S111, WARS1, PGM1, PGR, TNFRSF1B, TCF3, SURF1, SRY, SPARC, ACTB, PRB4, SNRPN, PRS, PXMP2, SGCA, REN, RFC1, PER2, MYO1A, FUS, SNURF, GBA, SORT1, RER1, GUSBP14, GBAP1, DCTN2, FBLN5, GJB2, GJB3, GLC3B, BCL2L11, GOT2, SCO2, HCLS1, HK1, HPT, IL2RB, KARS1, NRG2, STMN1, LCAT, ARHGEF2, MBP, GUSBP1
-
Glossitis
Wikipedia
Burning sensation. [2] Some use the term secondary burning mouth syndrome in cases where a detectable cause, such as glossitis, for an oral burning sensation. [5] Depending upon the underlying cause, there may be additional signs and symptoms such as pallor , oral ulceration and angular cheilitis. [2] Causes [ edit ] Anemias [ edit ] Iron-deficiency anemia is mainly caused by blood loss, such as may occur during menses or gastrointestinal hemorrhage . ... Alcoholism [7] Sprue ( celiac disease , [11] or tropical sprue ), secondary to nutritional deficiencies [7] Crohn’s disease [7] Whipple disease [7] Glucagonoma syndrome [7] Cowden disease [7] Acquired immunodeficiency syndrome (AIDS) [7] Carcinoid syndrome [7] Kwashiorkor amyloidosis [7] Veganism and other specialized diets , [2] Poor hydration and low saliva in the mouth, which allows bacteria to grow more readily Mechanical irritation or injury from burns, rough edges of teeth or dental appliances, or other trauma Tongue piercing [12] Glossitis can be caused by the constant irritation by the ornament and by colonization of Candida albicans in site and on the ornament [13] Exposure to irritants such as tobacco , alcohol , hot foods, or spices Allergic reaction to toothpaste , mouthwash , breath fresheners, dyes in confectionery, plastic in dentures or retainers, or certain blood-pressure medications ( ACE inhibitors ) Administration of ganglion blockers (e.g., Tubocurarine , Mecamylamine ). ... "Interventions for treating burning mouth syndrome" . The Cochrane Database of Systematic Reviews . 11 : CD002779. doi : 10.1002/14651858.CD002779.pub3 . ... PMID 15835487 . ^ Park AH, Batchra N, Rowley A, Hotaling A (May 1997). "Patterns of Kawasaki syndrome presentation". Int. J. Pediatr. ... "Prolonged use of a diaphragm and toxic shock syndrome". Fertil. Steril . 38 (2): 248–50. doi : 10.1016/s0015-0282(16)46467-8 .
-
Cyld Cutaneous Syndrome
Gene_reviews
Offspring of an individual with CYLD cutaneous syndrome have a 50% chance of inheriting the variant. ... Diagnosis Formal diagnostic criteria for CYLD cutaneous syndrome (CCS) have not been established. ... Individuals with CYLD cutaneous syndrome may present with more than one tumor type discussed below. ... Outdated terms previously used in the literature to refer to CYLD cutaneous syndrome include the following: Ancell-Spiegler cylindromas Dermal eccrine cylindroma Turban tumor syndrome (now denoted as confluent scalp tumors) Prevalence The true prevalence of CYLD cutaneous syndrome is unknown but may be in the order of more than 1:100,000 population [Rajan & Ashworth 2015]. ... Disorders with Multiple Facial Papules in the Differential Diagnosis of CYLD Cutaneous Syndrome (CCS) View in own window Gene(s) Disorder 1 Distinguishing Histologic Features in Differential Disorder Comment FLCN Birt-Hogg-Dubé syndrome Fibrofolliculomas NF1 Neurofibromatosis 1 (NF1) Neurofibromas Both NF1 & CCS are assoc w/lesions on the torso PTEN Cowden syndrome (see PTEN H amartoma Tumor Syndrome) Trichilemmomas TSC1 TSC2 Tuberous sclerosis complex Angiofibromas HR Marie Unna hypotrichosis 1 (MUHH1) (OMIM 146550) Trichoepitheliomas 2 Severe hair breakage/loss & absence of cylindromas in MUHH1 further distinguishes MUHH1 from CCS.
-
Cancer Syndrome
Wikipedia
Many cancer syndromes are inherited in this manner. ... If both parents have one mutant allele and one normal allele ( heterozygous ) then they have a 25% chance of producing a homozygous recessive child (has predisposition), 50% chance of producing a heterozygous child (carrier of the faulty gene) and 25% chance of produced a child with two normal alleles. [8] Examples of autosomal dominant cancer syndromes are autoimmune lymphoproliferative syndrome (Canale-Smith syndrome), Beckwith–Wiedemann syndrome (although 85% of cases are sporadic), [ citation needed ] Birt–Hogg–Dubé syndrome , Carney syndrome , familial chordoma , Cowden syndrome , dysplastic nevus syndrome with familial melanoma , familial adenomatous polyposis , hereditary breast–ovarian cancer syndrome , hereditary diffuse gastric cancer (HDGC), Hereditary nonpolyposis colorectal cancer (Lynch syndrome), Howel–Evans syndrome of esophageal cancer with tylosis , juvenile polyposis syndrome , Li–Fraumeni syndrome , multiple endocrine neoplasia type 1/2, multiple osteochondromatosis , neurofibromatosis type 1/2, nevoid basal-cell carcinoma syndrome (Gorlin syndrome), Peutz–Jeghers syndrome , familial prostate cancer , hereditary leiomyomatosis renal cell cancer (LRCC), hereditary papillary renal cell cancer , hereditary paraganglioma -pheochromocytoma syndrome, retinoblastoma , tuberous sclerosis , von Hippel–Lindau disease and Wilm's tumor . [9] Examples of autosomal recessive cancer syndromes are ataxia–telangiectasia , Bloom syndrome , Fanconi anemia , MUTYH-associated polyposis, Rothmund–Thomson syndrome , Werner syndrome and Xeroderma pigmentosum . [9] Examples [ edit ] Although cancer syndromes exhibit an increased risk of cancer, the risk varies. ... Tumors with increased risk in this disorder are colorectal cancer, gastric adenomas and duodenal adenomas. [15] [28] Micrograph showing keratocystic odontogenic tumour , a common finding in nevoid basal cell carcinoma syndrome. H&E stain . Nevoid basal cell carcinoma syndrome [ edit ] Nevoid basal cell carcinoma syndrome , also known as Gorlin syndrome, is an autosomal dominant cancer syndrome in which the risk of basal cell carcinoma is very high. ... PMID 15050066 . ^ Lo Muzio L (2008). "Nevoid basal cell carcinoma syndrome (Gorlin syndrome)" . Orphanet Journal of Rare Diseases . 3 : 32. doi : 10.1186/1750-1172-3-32 . ... PMID 19078925 . ^ Carethers JM, Stoffel EM (2015). "Lynch syndrome and Lynch syndrome mimics: The growing complex landscape of hereditary colon cancer" .RET, VHL, BAP1, TP53, PTEN, BRCA1, FH, MLH1, BRCA2, CDH1, MSH2, STK11, DICER1, TSC2, SDHB, PMS2, MSH6, APC, BMPR1A, CDKN2A, SDHAF2, MRPS18C, SMARCB1, RAD51B, RAD51D, RB1, CDKN2B-AS1, RECQL, FLCN, SDHA, SDHC, SDHD, SMARCA4, C11orf65, SMARCE1, TOE1, ABRAXAS1, BRIP1, XRCC2, FBXO11, PALB2, RAD50, RAD51C, CHEK2, MPRIP, TMEM127, HOXB13, POLE, PTCH1, MUTYH, BARD1, BLM, CDK4, CTNNA1, EXT2, FANCC, HFE, SMAD4, MAX, ATM, MITF, MRE11, TH2LCRR, NBN, NF1, POLD1, NTHL1, MRC1, MEN1, CDC73, CYP19A1, ST8, PLAU, BRAF, RUNX1, PKM, PIK3CG, CTNNB1, SORD, SDS, SARDH, PIK3CD, MSH3, PART1, PIK3CA, KRAS, SERPINE1, PIK3CB
-
Wolff–parkinson–white Syndrome
Wikipedia
Wolff–Parkinson–White syndrome Other names WPW pattern, Ventricular pre-excitation with arrhythmia, auriculoventricular accessory pathway syndrome [1] [2] A characteristic "delta wave" (arrow) seen in a person with Wolff–Parkinson–White syndrome. ... "The prevalence of the Wolff–Parkinson–White syndrome in a population of 116,542 young males". ... PMID 11526369 . ^ John Kenyon. Wolff–Parkinson–White Syndrome and the Risk of Sudden Cardiac Death. ... Available at: "Wolff-Parkinson-White Syndrome and the Risk of Sudden Cardiac Death" . ... "Atrial fibrillation in the Wolff–Parkinson–White syndrome: ECG recognition and treatment in the ED".
-
Myotonic Dystrophy
Wikipedia
History [ edit ] Myotonic dystrophy was first described by a German physician, Hans Gustav Wilhelm Steinert , who first published a series of 6 cases of the condition in 1909. [27] Isolated case reports of myotonia had been published previously, including reports by Frederick Eustace Batten and Hans Curschmann, and Type 1 myotonic dystrophy is therefore sometimes known as Curschmann-Batten-Steinert syndrome. [28] The underlying cause of type 1 Myotonic dystrophy was determined in 1992. [2] References [ edit ] ^ a b c d e f g h i j k l m n o p q r s t u v w x "myotonic dystrophy" . ... "[Case of Curschmann-Batten-Steinert syndrome]". Wiad. Lek. (in Polish). 34 (17): 1467–9. ... External links [ edit ] Myotonic dystrophy at Curlie Classification D ICD - 10 : G71.1 OMIM : 160900 MeSH : D009223 DiseasesDB : 8739 External resources GeneReviews : Myotonic Dystrophy Type 1 Orphanet : 206647 v t e Muscular dystrophy Types Congenital Dystrophinopathy Becker's Duchenne Distal Emery-Dreifuss Facioscapulohumeral Limb-girdle muscular dystrophy Calpainopathy Myotonic Oculopharyngeal National/International Organizations Muscular Dystrophy Association (USA) Muscular Dystrophy Canada Myotonic Dystrophy Foundation Muskelsvindfonden (Denmark) National/International Events MDA Muscle Walk (USA) Labor Day Telethon (defunct; USA/Canada) Décrypthon (France) Grøn Koncert (Denmark) Clinical trials Stamulumab (MYO-029) Category v t e Diseases of muscle , neuromuscular junction , and neuromuscular disease Neuromuscular- junction disease autoimmune Myasthenia gravis Lambert–Eaton myasthenic syndrome Neuromyotonia Myopathy Muscular dystrophy ( DAPC ) AD Limb-girdle muscular dystrophy 1 Oculopharyngeal Facioscapulohumeral Myotonic Distal (most) AR Calpainopathy Limb-girdle muscular dystrophy 2 Congenital Fukuyama Ullrich Walker–Warburg XR dystrophin Becker's Duchenne Emery–Dreifuss Other structural collagen disease Bethlem myopathy PTP disease X-linked MTM adaptor protein disease BIN1-linked centronuclear myopathy cytoskeleton disease Nemaline myopathy Zaspopathy Channelopathy Myotonia Myotonia congenita Thomsen disease Neuromyotonia / Isaacs syndrome Paramyotonia congenita Periodic paralysis Hypokalemic Thyrotoxic Hyperkalemic Other Central core disease Mitochondrial myopathy MELAS MERRF KSS PEO General Inflammatory myopathy Congenital myopathy v t e Non-Mendelian inheritance : anticipation Trinucleotide Polyglutamine (PolyQ), CAG Dentatorubral-pallidoluysian atrophy Huntington's disease Kennedy disease Spinocerebellar ataxia 1, 2, 3, 6, 7, 17 ( Machado-Joseph disease ) Non-polyglutamine CGG ( Fragile X syndrome ) GAA ( Friedreich's ataxia ) CTG ( Myotonic dystrophy type 1 ) CTG ( Spinocerebellar ataxia 8 ) CAG ( Spinocerebellar ataxia 12 ) Tetranucleotide CCTG ( Myotonic dystrophy type 2 ) Pentanucleotide ATTCT ( Spinocerebellar ataxia 10 ) v t e Deficiencies of intracellular signaling peptides and proteins GTP-binding protein regulators GTPase-activating protein Neurofibromatosis type I Watson syndrome Tuberous sclerosis Guanine nucleotide exchange factor Marinesco–Sjögren syndrome Aarskog–Scott syndrome Juvenile primary lateral sclerosis X-Linked mental retardation 1 G protein Heterotrimeic cAMP / GNAS1 : Pseudopseudohypoparathyroidism Progressive osseous heteroplasia Pseudohypoparathyroidism Albright's hereditary osteodystrophy McCune–Albright syndrome CGL 2 Monomeric RAS: HRAS Costello syndrome KRAS Noonan syndrome 3 KRAS Cardiofaciocutaneous syndrome RAB: RAB7 Charcot–Marie–Tooth disease RAB23 Carpenter syndrome RAB27 Griscelli syndrome type 2 RHO: RAC2 Neutrophil immunodeficiency syndrome ARF : SAR1B Chylomicron retention disease ARL13B Joubert syndrome 8 ARL6 Bardet–Biedl syndrome 3 MAP kinase Cardiofaciocutaneous syndrome Other kinase / phosphatase Tyrosine kinase BTK X-linked agammaglobulinemia ZAP70 ZAP70 deficiency Serine/threonine kinase RPS6KA3 Coffin-Lowry syndrome CHEK2 Li-Fraumeni syndrome 2 IKBKG Incontinentia pigmenti STK11 Peutz–Jeghers syndrome DMPK Myotonic dystrophy 1 ATR Seckel syndrome 1 GRK1 Oguchi disease 2 WNK4 / WNK1 Pseudohypoaldosteronism 2 Tyrosine phosphatase PTEN Bannayan–Riley–Ruvalcaba syndrome Lhermitte–Duclos disease Cowden syndrome Proteus-like syndrome MTM1 X-linked myotubular myopathy PTPN11 Noonan syndrome 1 LEOPARD syndrome Metachondromatosis Signal transducing adaptor proteins EDARADD EDARADD Hypohidrotic ectodermal dysplasia SH3BP2 Cherubism LDB3 Zaspopathy Other NF2 Neurofibromatosis type II NOTCH3 CADASIL PRKAR1A Carney complex PRKAG2 Wolff–Parkinson–White syndrome PRKCSH PRKCSH Polycystic liver disease XIAP XIAP2 See also intracellular signaling peptides and proteins v t e Genetic disorders relating to deficiencies of transcription factor or coregulators (1) Basic domains 1.2 Feingold syndrome Saethre–Chotzen syndrome 1.3 Tietz syndrome (2) Zinc finger DNA-binding domains 2.1 ( Intracellular receptor ): Thyroid hormone resistance Androgen insensitivity syndrome PAIS MAIS CAIS Kennedy's disease PHA1AD pseudohypoaldosteronism Estrogen insensitivity syndrome X-linked adrenal hypoplasia congenita MODY 1 Familial partial lipodystrophy 3 SF1 XY gonadal dysgenesis 2.2 Barakat syndrome Tricho–rhino–phalangeal syndrome 2.3 Greig cephalopolysyndactyly syndrome / Pallister–Hall syndrome Denys–Drash syndrome Duane-radial ray syndrome MODY 7 MRX 89 Townes–Brocks syndrome Acrocallosal syndrome Myotonic dystrophy 2 2.5 Autoimmune polyendocrine syndrome type 1 (3) Helix-turn-helix domains 3.1 ARX Ohtahara syndrome Lissencephaly X2 MNX1 Currarino syndrome HOXD13 SPD1 synpolydactyly PDX1 MODY 4 LMX1B Nail–patella syndrome MSX1 Tooth and nail syndrome OFC5 PITX2 Axenfeld syndrome 1 POU4F3 DFNA15 POU3F4 DFNX2 ZEB1 Posterior polymorphous corneal dystrophy Fuchs' dystrophy 3 ZEB2 Mowat–Wilson syndrome 3.2 PAX2 Papillorenal syndrome PAX3 Waardenburg syndrome 1&3 PAX4 MODY 9 PAX6 Gillespie syndrome Coloboma of optic nerve PAX8 Congenital hypothyroidism 2 PAX9 STHAG3 3.3 FOXC1 Axenfeld syndrome 3 Iridogoniodysgenesis, dominant type FOXC2 Lymphedema–distichiasis syndrome FOXE1 Bamforth–Lazarus syndrome FOXE3 Anterior segment mesenchymal dysgenesis FOXF1 ACD/MPV FOXI1 Enlarged vestibular aqueduct FOXL2 Premature ovarian failure 3 FOXP3 IPEX 3.5 IRF6 Van der Woude syndrome Popliteal pterygium syndrome (4) β-Scaffold factors with minor groove contacts 4.2 Hyperimmunoglobulin E syndrome 4.3 Holt–Oram syndrome Li–Fraumeni syndrome Ulnar–mammary syndrome 4.7 Campomelic dysplasia MODY 3 MODY 5 SF1 SRY XY gonadal dysgenesis Premature ovarian failure 7 SOX10 Waardenburg syndrome 4c Yemenite deaf-blind hypopigmentation syndrome 4.11 Cleidocranial dysostosis (0) Other transcription factors 0.6 Kabuki syndrome Ungrouped TCF4 Pitt–Hopkins syndrome ZFP57 TNDM1 TP63 Rapp–Hodgkin syndrome / Hay–Wells syndrome / Ectrodactyly–ectodermal dysplasia–cleft syndrome 3 / Limb–mammary syndrome / OFC8 Transcription coregulators Coactivator: CREBBP Rubinstein–Taybi syndrome Corepressor: HR ( Atrichia with papular lesions )DMPK, CNBP, NKX2-5, CELF1, MBNL1, CCT3, APOC2, INSR, SIX5, CKM, ERCC1, CLCN1, FXN, RANGAP1, PRKCA, HCRT, PRRT2, DMD, BUB1, PRKCB, POMC, NEK6, DMWD, MAPT, ACTB, CEBPD, MBNL2, SCN4A, TNF, QPCT, KCNN3, TNNI3, IGHD1-7, MSH3, UGT8, OXA1L, PLCB1, PGD, SCN5A, CDC42BPB, ALB, RAN, REM1, TNNT1, GH1, FSD1L, FSD1, BCL3, APOC1, MIR206, BIN1, GSK3B, CLIP2, MAK16, GGTLC1, RIDA, BPIFA2, OPN1MW3, OPN1MW2, SLC35G1, ROCK2, WASL, MTMR1, C9orf72, CTCF, CDC42BPA, DYSF, ST8SIA4, MIR29C, CELF2, RAB6B, FAM107B, ASB2, TNFRSF12A, TP53, RBFOX1, NAT10, IGHD1-14, DESI1, RBMS3, PDLIM3, FGF21, PNO1, ATRNL1, JPH3, MMD, CELF6, SRRM2, GGCT, ASRGL1, SPEN, LDB3, RSPH6A, RAB1B, CCL27, RAP2B, TCF4, CYP2B6, KCNQ1, IL6, IGLC3, GPR4, GHRH, OPN1MW, GC, FRAXE, FEN1, F5, DDX6, DDX5, CYP2A13, MEF2A, CYP2A7, CYP2A6, CDC42, CD59, CCND3, ATP1A3, ATHS, ARF3, APOE, AMPH, AMH, ALPP, LDLR, MEF2C, SYN1, PSPH, STXBP3, SRF, SLPI, SLC1A2, SRSF2, ATXN1, CLIP1, RRAS, ROCK1, REG1A, RAP1B, PTBP1, PSPN, ATXN3, PRNP, PRKCG, PRKAB1, PRKAA2, PRKAA1, PEPD, MYOG, MTM1, MSMB, MSH2, MRC1, MLH1, LOC102724197
-
Hereditary Diffuse Gastric Cancer
Gene_reviews
Variants in TNF and IFNGR1 can significantly increase the risk for gastric cancer, particularly in those infected with virulent strains of H pylori [Canedo et al 2008]. Other Cancer Predisposition Syndromes Gastric cancer is seen in several other autosomal dominant cancer predisposition syndromes, including Lynch syndrome (hereditary non-polyposis colorectal cancer), Li-Fraumeni syndrome, familial adenomatous polyposis, Peutz-Jeghers syndrome, and Cowden syndrome (one of the PTEN hamartoma tumor syndrome phenotypes). Lynch syndrome , which is associated with germline pathogenic variants in mismatch repair genes or EPCAM , predisposes heterozygotes to colorectal and other cancers. ... IGC is the predominant subtype in Lynch syndrome [Lynch et al 2005, Capelle et al 2010]. ... The incidence of gastric cancer is significantly higher in individuals with FAP in the East Asian population [Yamaguchi et al 2016]. Li-Fraumeni syndrome (LFS). Cancers in LFS are caused by pathogenic variants in TP53 . ... All individuals have immediate as well as long-term complications including rapid intestinal transit, dumping syndrome, diarrhea, eating habit alterations, and weight loss [Caldas et al 1999, Lewis et al 2001].
-
Endometrial Cancer
Wikipedia
Overall, hereditary causes contribute to 2–10% of endometrial cancer cases. [3] [24] Lynch syndrome , an autosomal dominant genetic disorder that mainly causes colorectal cancer , also causes endometrial cancer, especially before menopause. Women with Lynch syndrome have a 40–60% risk of developing endometrial cancer, higher than their risk of developing colorectal (bowel) or ovarian cancer. [17] Ovarian and endometrial cancer develop simultaneously in 20% of people. Endometrial cancer nearly always develops before colon cancer, on average, 11 years before. [18] Carcinogenesis in Lynch syndrome comes from a mutation in MLH1 or MLH2 : genes that participate in the process of mismatch repair , which allows a cell to correct mistakes in the DNA. [17] Other genes mutated in Lynch syndrome include MSH2 , MSH6 , and PMS2 , which are also mismatch repair genes. Women with Lynch syndrome represent 2–3% of endometrial cancer cases; some sources place this as high as 5%. [18] [21] Depending on the gene mutation, women with Lynch syndrome have different risks of endometrial cancer. ... There is an apparent link with these genes but it is attributable to the use of tamoxifen, a drug that itself can cause endometrial cancer, in breast and ovarian cancers. [17] The inherited genetic condition Cowden syndrome can also cause endometrial cancer.PTEN, ESR1, FGFR2, PGR, VEGFA, CCND1, ESR2, CDH1, MSH6, EP300, CYP11A1, MYC, STAR, SRD5A2, AKR1C1, HOXA10, AKR1C3, TNFSF10, SPOP, HOXA11, ABCC9, SOCS3, SRSF10, DCN, EZH2, ARID1A, SEMA3B, HSD17B2, NRIP1, SEMA3F, SIX1, POLD1, PAWR, NFE2L2, SULT2B1, MTHFR, MSH3, ZEB1, MAP3K4, TP53, LASP1, CXCL8, MLH3, IGF1R, SUZ12, CCL2, CHD4, BIRC5, TSPYL2, RNF43, PLXNA3, CDKN1C, JAZF1, FBXW7, AKR1B10, RNF144B, AKR1B1, CHFR, MIR200C, CDKN2A, MET, STK11, ERRFI1, CDKN2B, CTNNB1, HNF1B, LSAMP, SH2B3, RNGTT, FAM182A, ATXN2, ECT2L, RETREG3, TMTC1, SCAF8, CLEC3A, CYP19A1, TDP2, PTGS2, MLH1, MSH2, PIK3CA, AR, MMP2, KRAS, CYP17A1, BCL2, CYP1B1, AKT1, CCN1, ERBB2, MMP9, CFLAR, COMT, STS, EMP2, FAS, SULT1A1, CD82, PPIA, SULT1E1, SERPINB2, MGMT, MDM2, LEPR, FOXO1, TNF, MMP26, WT1, IL6, EBAG9, HIF1A, IL1A, NR1I2, CTNNBIP1, RDH16, SLC12A9, CHEK2, HOXB13, FST, NDRG1, SREBF1, PROM1, PROK1, SPP1, ACKR3, SIGLEC1, SKP2, PELP1, AURKB, SHBG, ADIPOQ, SIRT1, NAPSA, SDC1, CASC2, TCF7L2, RASSF1, UGT1A1, TTF1, WNT7A, TSC2, HSP90B1, SLC22A16, HTRA3, HPSE, LRIG2, TLR4, TRIM25, FSTL1, SCGB1A1, TLR3, CXCR4, NKX2-1, C19orf33, TFF3, NCOA3, TERT, ARMC3, KIAA1324, IGF2BP3, ADM, MMP13, S100A8, CLDN4, CRP, CSNK2B, VCAN, CXADR, CYP1A1, CYP3A4, DNMT1, TYMP, EGFR, ENG, EPO, EPOR, ERCC1, ETS1, ETV5, F2RL1, FABP5, FHIT, FOS, MTOR, NR5A2, CLDN3, CPE, S100A4, MAP3K8, NR0B1, ALDH1A1, APC, BIRC2, ATM, CEACAM1, BMI1, BRCA1, BRAF, BRCA2, DDR1, CAV1, RUNX3, CCND3, CD44, CDC25B, CDH13, CDK6, CDKN1A, CDX2, CLU, NR5A1, GH1, GJB2, GSTP1, SCGB2A1, MME, MMP7, ADRB3, MMP14, MSI1, COX1, MUTYH, NEDD4, NFKB1, OVGP1, PCNA, SERPINF1, ABCB1, SERPINB5, PIK3R1, PLAU, PPARG, PRKCA, HTRA1, RARB, MDK, SMAD7, LLGL1, HSPE1, HAS1, HDAC1, HMMR, NR4A1, FOXA1, SLC29A2, HRAS, HSD3B2, HSD17B1, IGF1, L1CAM, IGFBP1, IL1B, IL1RN, INHBA, CXCL10, ITGB1, JUN, JUP, KCNN4, PERCC1
-
Breast Cancer
Omim
Breast cancer is a feature of several cancer syndromes, including Li-Fraumeni syndrome (151623) due to germline mutations in p53; Cowden syndrome (158350) due to mutations in the PTEN gene (601728); and Peutz-Jeghers syndrome (175200) due to mutations in the STK11 gene (602216). ... Soft tissue sarcomas are associated with breast cancer in Li-Fraumeni syndrome. Mulvihill (1982) used the term cancer family syndrome of Lynch (120435) for the association of colon and endometrial carcinoma and other neoplasms including breast cancer. ... Other Features Chang et al. (1987) showed that the noncancerous skin fibroblasts of members of a family with Li-Fraumeni syndrome (which show resistance to the killing effect of ionizing radiation) have a 3- to 8-fold elevation in expression of the MYC oncogene (190080) and an apparent activation of the RAF1 gene (164760). ... Lynch et al. (1984) found evidence consistent with a hereditary breast cancer syndrome in 5% of 225 consecutively ascertained patients with verified breast cancer. ... Kainu et al. (2000) adopted a strategy similar to that used in the identification of the locus for the Peutz-Jeghers cancer syndrome (175200), based on the Knudson 2-hit model of development: detection of somatic deletions in the wildtype gene by comparative genomic hybridization (CGH) followed by targeted linkage analysis.PALB2, XRCC2, RAD51D, ATM, BRCA1, BRCA2, CHEK2, BARD1, RAD50, NBN, SLX4, MLH1, MRE11, MUTYH, RAD51C, CHEK1, RECQL, PPM1D, MSH6, CDH1, MSH2, MCPH1, RAD54L, KLLN, GEN1, SLC22A18, PIK3CA, PMS2, RINT1, EPCAM, RAD51, RET, TP53, BRIP1, ERBB2, ESR1, PTEN, BRCA3, FANCM, PARP1, COL11A2, XRCC1, ARL11, MDM2, RASSF1, NLRP2, TSG101, STK11, GADD45A, PHB, IGF1, HSD17B2, CYP17A1, BACH1, BLM, EMSY, TCF7L2, MGMT, CYP1A1, PRL, FANCI, MYLIP, RTEL1, MTUS1, UIMC1, VEGFA, MARCHF8, GSTP1, FANCD2, AKAP13, MLXIP, FGFR2, HSD17B1, CD44, WRN, CASP8, POLQ, BRD7, PPARGC1A, TOPBP1, DICER1, WIF1, FOXP1, MAST1, BABAM1, AKAP10, AATF, KLK14, FOXD3, TOX3, HPGDS, RHOBTB2, FANCB, CT55, KRT88P, MIR1179, ANGPT2, CD24, MIR499A, MIR17HG, MIR30C2, MIR30C1, MIR27A, MIR21, MIR17, APC, PPARGC1B, AR, FANCL, ABRAXAS1, ATR, PIF1, CCND1, ARHGEF28, CLSPN, BCORL1, BUB1B, VPS51, SH3RF1, CASP10, TEX15, LHFPL6, BRAP, SAFB2, RECQL5, NFKB1, CYP19A1, MYC, MTHFR, MSR1, MSH3, EGFR, EP300, KMT2A, ERCC4, MDM4, FANCA, LSS, KRAS, JAK2, CXCL10, IGFBP3, HTC2, HRAS, TLX1, HMGA1, HIF1A, GSTT1, GSTM1, SFN, FOXC1, FHIT, PER1, ABCB1, CYP2D6, CDKN2A, SLC9A3R1, PDLIM7, EXO1, CLDN1, PER2, LMO4, CAV1, CCND2, XRCC3, UCHL1, TTK, TPD52, TM7SF2, POLH, TGFBR1, TEK, SULT1A1, SOAT1, SAFB, CENPE, ANGPT1, RCC1, RAD51B, COMT, PTPRG, CTNNB1, PSC
-
Papillary Thyroid Cancer
Wikipedia
Environmental exposures to radiation such as atomic bombings of Hiroshima and Nagasaki and Chernobyl disaster also causes an increase in childhood papillary thyroid cancer at 5 to 20 years after the exposure to radiation. [38] Family history of thyroid cancer syndrome such as familial adenomatous polyposis , Carney complex , Multiple endocrine neoplasia type 2 (MEN-2), Werner syndrome , and Cowden syndrome increases the risk of getting papillary cancer. [37] References [ edit ] ^ a b c d e f Chapter 20 in: Mitchell, Richard Sheppard; Kumar, Vinay; Abbas, Abul K; Fausto, Nelson (2007).BRAF, NKX2-1, CCDC6, RET, FOXE1, NCOA4, PPARG, KRAS, PAX8, NTRK1, NRAS, DIRC3, NTRK3, ETV6, ALK, GAS8-AS1, PTCSC3, HRAS, MIR485, EIF1AX, SNAI1, ERC1, TPR, TFG, DIO3, MIR431, MIR381, MIR409, MIR369, MIR376C, MIR758, PCM1, TRIM27, MIR299, MIR539, MIR136, TRIM33, LPAR4, MIR654, TRIM24, MIR379, MIR382, WARS1, WDR20, MIR380, BEGAIN, NDUFA13, MIR377, MIR376A1, MIR127, MIR370, MIR134, MIR337, MIR323A, MIR433, MIR154, DIO3OS, MIR329-1, MIR496, MIR323B, MIR376A2, MIR487B, MIR411, MIR655, MIR656, MIR770, MIR300, MIR543, DYNC1H1, MIR889, MIR1247, MIR1197, MIR1193, ZEB1, GOLGA5, MIR487A, MIR493, MIR410, PPP2R5C, MIR412, MIR432, MIR494, SNAI2, MIR376B, ZEB2, MIR495, APC, HABP2, PTEN, NRG1, DICER1, TAS2R38, PTCH1, MINPP1, MAPK1, PIK3CG, TPCN1, PIK3CD, PIK3CB, CTNNB1, PIK3CA, PCNX2, ESR1, EGFR, F9, VAV3, TP53, BRCA2, MIR146B, BCL2, CCND1, TGFB1, HT, AKT1, JAG1, TG, LINC02454, TERT, LOC110806263, LGALS3, MIR222, SEC23B, MMP9, CXCR4, LIG4, TPM3, TPO, TSHR, SLC5A5, MIR221, VEGFA, EPHB2, MIR146A, COX2, FN1, CD274, MET, KRT19, MIR21, PTGS2, TIMP1, GABPA, SPP1, NCAM1, MTCO2P12, ATM, MUC1, MAP2K7, IGF1, KIT, TNF, ESR2, CD44, HIF1A, RASSF1, SLC2A1, CXCL12, HGF, VEGFC, NFE2L2, MAPK3, TTF1, MIR204, PRPF31, VDR, DUSP6, CDKN1B, MIR451A, XIAP, TFF3, PDGFRA, CYP24A1, THBS1, KDR, RUNX2, PTH, LEP, CHEK2, STAT3, ERBB2, MMP2, MIR144, PCNA, RAF1, VIM, CDKN2A, PTCH2, MDK, IRAK1, MTOR, MT1G, ROCK1, SOX2, IGF1R, CITED1, S100A4, GDF1, MYC, NOTCH1, LEPR, MDM2, TACSTD2, H3P10, ICAM1, FOXP3, MIR30A, CRK, ACKR3, TGFA, NOB1, NEAT1, MIR141, SMUG1, MIR155, MIR15A, HTC2, AKT3, DPP4, SPHK1, CENPJ, UCA1, HMGA2, TIMP3, EDN1, MIR486-1, XRCC1, AR, SNHG12, MIR613, IL6, SLC5A8, PTCSC2, MT1A, MMP11, WNK1, ORI6, SPHK2, MALAT1, MIR34B, IL10, MIR150, MIR96, MLH1, MIR199A1, MIR199A2, MCL1, MIR200A, SYTL2, MIR497, MIR29A, MIR206, SP1, KRT20, AIMP2, AHSA1, GRAP2, RAP1A, TP63, TTF2, RNH1, RPE65, KLK7, S100A11, WNT5A, VHL, UVRAG, SLC2A3, TWIST1, PPP1R13L, PROX1, TERF2IP, SGSM3, SHC3, GDE1, SIRT6, IL22, STOML2, PDGFRB, RABGEF1, CKAP4, PDCD4, POLDIP2, RNF19A, DKK1, ZHX2, MAP2K1, YAP1, ABL1, MS4A1, COL1A1, VEGFD, CCR7, HLA-G, MAPK14, DIO2, CDH1, HLA-DRB1, DIO1, EZH2, HOXD13, EDNRA, FOXO1, FAS, EGF, HMGB1, GSTM1, GPI, HSP90AA1, MIR122, MIR139, DCTN6, EBP, SOX11, GLI1, NNMT, TNFSF10, MIR202, MIR137, MIRLET7E, CDKN1A, NRP1, HOPX, TENM1, ITCH, PLAU, AKAP9, PLG, EIF4E, SLPI, DNMT1, SERPINA1, PGR, DDR2, NRCAM, P2RX7, MIR126, PAK1, ZNRD2, FGF2, PDPN, NT5E, CIB1, PCBP4, POSTN, FGFR1, CRNDE, FASLG, POLR2E, ZNRF3, KIDINS220, NOTCH3, MIR10A, PDPK1, ARNTL, MIR509-1, MIR149, MIR145, DUSP5, RAC1, CAV1, MIR7-2, RARA, MIR34A, RARG, NAPSA, MIR195, OPN1LW, RELA, CCK, ABCG2, SLIT2, MIR363, FOS, FLT3, CCL21, FLT1, RPL36A, CLDN10, MIR199B, CLDN1, S100A6, FOXO3, TMSB10, LPAR2, MIAT, MIR183, CXCL14, PTN, CDK4, PAG1, PRKAA1, PRKAA2, PRKAB1, DNMT3A, CA12, GAS5, FOXM1, MIR375, GAS1, CDH6, KEAP1, CASP3, EIF4A3, RAPGEF5, GORASP1, MIR335, PSMD9, E2F1, SFTPB, GABRB2, NECTIN4, SELL, FRTS1, NCL, C3, SYT1, TOP2A, TRAF6, JAK1, JUN, CXXC5, IL27, CASC2, HOXA@, LAMB3, LAMC2, IL17D, CREBBP, ITGA3, LINC00313, HAGLR, LGALS1, CPSF2, LMNA, MIR2861, TLR4, ERRFI1, LRP4, CRABP1, ITGA5, ITGA2, SIRT1, IL1B, H3P23, YY1, CLIP2, KLB, IFI27, DEUP1, IGFBP5, CTSC, IHH, IL1A, ZCCHC12, FALEC, HSPA5, CXCL8, CXCR2, ABHD11-AS1, LIPH, IL13RA2, IL17A, TMED7, CXCL10, INSR, EPCAM, LPAR3, ANGPT1, MTDH, PTK2B, TMED10, MT1X, MIR933, AHR, AKT2, CISH, TMED7-TICAM2, TMED10P1, GSTT1, TICAM2, TEK, ALDH1A1, MST1, DCN, MSH2, FOXD2-AS1, UNC5B-AS1, HOTAIR, HLA-C, HOXA-AS2, MCM5, CYP27B1, ETV5, MDM4, HLA-DQB1, CYTOR, THRB, RASAL1, KDM1A, KLLN, PDCD1LG2, NT5C1A, STON2, DCSTAMP, SLCO6A1, PROK1, GPR151, ADAT2, E2F7, PROSER3, CLPTM1L, AFAP1L2, LIMD2, SPZ1, RTN4IP1, LMLN, CHEK1, TMEM139, FSD1L, ECRG4, XKR4, AHNAK2, FOXQ1, FAM83F, HTRA3, ABHD11, TMPRSS13, PKHD1L1, MCM3AP-AS1, SH3BGRL3, HSDL2, MAL2, ARL11, RBM45, MRGPRX3, TLR10, MRGPRX4, PRAP1, LINC01278, CYP2R1, ACBD5, CPT1C, TRIM8, HOOK3, KLF17, CHI3L1, HSPD1, WNT10A, IL17RB, LARP7, CRKL, RAB23, CREM, FXYD5, DUOX1, TLR9, CREB1, DLL4, CROT, SLC35F2, XAF1, TRIM44, GATAD2A, CPOX, PGPEP1, ELOVL2, PINX1, TUG1, PIWIL2, THAP1, RMDN3, ZNF654, LAPTM4B, IMPACT, CRY2, MBIP, IL23A, DUOX2, SETD2, DROSHA, PYCARD, CTLA4, UHRF1, TFCP2L1, CERS2, PSAT1, EHD2, CSNK1G2, CD207, NOX4, SIRT7, CSF2, TAS2R3, F11R, CSF1R, CSF1, RMDN1, HSPA14, TCEAL9, MZB1, TNFRSF12A, REV1, LGR4, SYBU, NAA15, IWS1, CXCL16, IL22RA1, ACE2, LGR6, SCOC, PROK2, MYO1G, ERAP2, SOX17, HHIP, MRPL41, CDK15, MRPL44, CCR3, TRAK2, OTUB2, MAPKAP1, FSD1, MUL1, TNFAIP8L2, LIN28A, E2F8, CKS2, TRPM3, ZNF703, C6orf47, ANKRD36B, NCOA5, TCIM, RUFY2, WDR11, ATF7IP, CHFR, AGK, COX8A, PBK, MYDGF, PDGFC, TMPRSS4, KLF6, EMSY, TNRC6C, SMYD2, CTNNBIP1, TWSG1, CEMIP, DANCR, SRGAP1, MIB1, SEMA6A, MRTFA, CNGB1, RELCH, TEKT4, MIR130A, RMDN2, GGTLC4P, MIR622, MIR625, MIR630, AMPD1, AMFR, MIR663A, LINC00460, GGTLC3, GGT2, CCR2, ALOX5, ALDH1A3, MIR608, MIR766, MIR675, ALCAM, VTRNA2-1, MIR744, MIR922, MIR940, MIR885, TNRC6C-AS1, LINC00271, CD24, ANGPT2, MIR599, MIR1915, ARAF, BMI1, BGN, BGLAP, BAX, B2M, ATP5F1E, ATF1, ARR3, MIR520A, MIR524, MIR506, RASSF10, MIR584, NORAD, CXADRP1, GGTLC5P, PAX8-AS1, APRT, APOA4, ANXA5, ANXA1, MIR564, MIR574, MIR577, MIR1183, NR0B1, CCDC80, LOC102723407, MIR4516, MIR4429, MIR5189, BANCR, ADRA2B, COMETT, NAMA, BLACAT1, SAMMSON, LINC01186, ADRA1A, LOC102724971, MIR4728, CCND2-AS1, BISPR, PARP1, CERNA3, LOC105379528, RNU1-55P, ADCYAP1R1, ADCYAP1, ADCY1, LNCRNA-ATB, H3P17, MIR4500, COMMD3-BMI1, MIR1270, MIR320E, MIR1266, MIR1271, MIR1261, MIR1179, MIR1304, AFM, MIR718, MIR761, HOTTIP, AP2A1, MIR4316, MIR3144, SNHG16, MIR3151, LINC00673, MIR3619, MIR3663, PROX1-AS1, APTR, LINC01672, LUCAT1, HOTAIRM1, TTN-AS1, DLG1-AS1, BMP4, BMPR1A, BRCA1, LINC01061, SCAI, UBAC2, RSPO2, ZNF677, GSTK1, AGRN, CDX2, KMT5A, CCL4L1, IYD, LOC390714, MIRLET7B, SNHG15, MIR106A, MIR106B, MIR125A, CDKN3, HIPK2, MIR130B, CDKN1C, CDK8, MIR143, CDK7, MIR148A, SLC26A4-AS1, RPL34-AS1, BRS3, USF3, PAQR3, C8orf37, CBLL2, OXER1, DACT2, GLIS3, NLRP6, EMX2OS, TIGIT, PRSS55, ANO5, SKA1, RSPO1, HOXA11-AS, JAZF1, GPRC6A, ZNRF2, TTTY10, FNDC5, MRGPRX1, MAGEA2B, CEBPB, B4GALNT3, LINC00514, CDK2, CDH2, MIR18A, MIR361, ZFAS1, GPR166P, MIR148B, CASR, MIR324, CASP9, MIR339, MIR346, CASP8, CASP6, MIR196B, CASP2, MIR181A2, CAMP, CALCA, CAD, CA9, MIR384, MIR422A, RGMB-AS1, MIR20B, TMEM50B, MPPED2, BUB1B, VN1R17P, NRARP, NR2F1-AS1, PRDM16-DT, MIR182, MIR190A, CD74, CD68, MIR20A, CD63, MIR203A, CD38, CD34, MIR211, MIR212, MIR214, MIR215, MIR219A1, MIR22, CD247, CD1A, MIR23A, CCNG1, MIR296, CBL, MIR31, RUNX3, ATAD2, AKAP13, CTNNA1, PTGDS, MAPK8, LRRC32, PRL, PRLR, GAPDH, GAP43, HTRA1, KLK10, PSG1, PSMD8, GAGE5, GAGE4, PTGIS, FLVCR1, GAGE1, G6PD, PTPRJ, NECTIN1, PVT1, RAD52, FUT4, FUCA1, RAP1GAP, RARB, RARRES2, RBP2, GCGR, GGT1, PRKCE, PRKAR1A, SLC26A4, PECAM1, PGM1, PHB, SERPINB5, SERPINE2, GCLC, GPC3, GLA, PIN1, PITX2, PKM, PLA2G1B, PLA2G2A, PLD2, PLK1, PMP22, PMS2, POMC, POU5F1B, PPARA, GJB2, GH1, PRKACA, PRKACG, FPR2, FOSB, RGS4, FKBP5, SLC6A9, SLC16A2, FKBP4, SOD2, SOD3, FGFR4, SOX9, SOX12, SPARC, SPG7, SPINT1, SRC, SRF, SRY, SST, SSTR4, STAT1, FGF1, STK11, STRN, SYPL1, MAP3K7, TBX1, TCF4, TBX3, SLC6A2, FOXF1, RNASE2, SLC1A5, BRD2, ROS1, RRAS, RXRG, S100A1, FLI1, S100A13, S100B, SERPINB3, SCD, SCN4B, CCL2, CCL3, CCL4, CCL5, CCL20, CXCL11, CXCL5, SDC4, SDHB, SELE, SFRP1, SHH, SHMT1, SKP2, ENPP2, GLI2, GLP1R, JUNB, CD82, KCNJ2, HOXD10, KIF5B, HOXA9, HOXA3, LAIR1, LASP1, LBR, LCN2, LDHA, HNF4A, FOXA2, HMGB2, LMO7, LOX, LOXL1, BCAM, HMBS, SMAD3, SMAD4, MAGEA2, MARK1, MC1R, MCM3, JUND, ITGB4, MAP3K5, ITGB2, TNC, HSP90AB1, ID1, ID3, IDH1, IFNG, HSPA9, IGFBP1, IGFBP7, IL1R1, IL2, IL2RA, IL4, HPRT1, IL11, IL11RA, IL13, IL18, IDO1, INS, INPPL1, IRS1, ISG20, ITGAV, ITGB1, MCM7, MEN1, PDCD1, GSN, GPX3, GRK6, NFKB1, NGF, NGFR, NNAT, NME1, NOS2, NOS3, GPR42, UTS2R, CXCR3, ROR2, NUCB2, PEBP1, PRDX1, SERPINE1, PRKN, PAX6, PC, SERPINA5, GNAS, GLS, CDK16, PCYT1A, MYH9, MMUT, MFAP1, MUC4, MGMT, CXCL9, MKI67, HLA-B, MME, MMP1, HLA-A, HK2, HIC1, MMP13, MOG, MPZ, MRC1, ABCC1, HBB, MSH3, MSMB, GUSB, MT1E, MT1F, MT1M, MT2A, MTF1, MTHFR, GSR, TCF7L2, FCGR3B, TERC, TAB1, CFD, ATG7, ARFGEF1, NPC2, TXNRD2, DECR1, PDLIM5, CXCR6, CLDN16, CERS1, POLQ, PTGES3, CHL1, FRS2, AKR1C2, HPSE, HCP5, EHD1, RAB40B, DCT, COPS6, OGFR, WDR5, ACE, HUS1, GPNMB, NQO1, ANKRD26, RAPGEF3, WSCD2, NUP93, RAB11FIP3, SART3, HDAC9, HDAC4, TRIM14, DPT, PJA2, TLK1, WDR1, FGF19, NR1I3, RBX1, SCO2, NAALADL1, PLXNC1, PSME3, TRIM13, SPRY2, STUB1, PAK4, TLR6, NDRG1, RACK1, TUSC2, CILK1, DUSP2, CYP1A1, LRIG1, DNM3, CYLD, CXADR, DGCR5, PHGDH, FGF21, EHF, BEX3, LAT, TSPAN13, DKK3, SNX5, SERP1, CUX1, HPGDS, RBMX, CTSL, CTSB, EML4, IGHV3OR16-7, IGHV3-69-1, TRBV2, TRBJ1-2, TRBJ1-1, WWTR1, BRMS1, DCK, QPCT, MMRN1, ATF6, RPIA, DAPK1, USP33, SMG1, RRS1, KAZN, CUX2, USP22, CYP19A1, ANGPTL2, HEY1, BRD4, PES1, SRRM2, PSD4, CYP2D6, AMACR, LDOC1, SNHG1, TRIM29, SLC7A11, IL17RA, RASGRP3, PRDX6, CCL4L2, FCGR3A, EPHB4, SUMO1, UGCG, USF1, EPHB1, VAV2, EPAS1, ENG, EMX2, ELK3, ELF3, TRPV1, ELAVL2, WIPF1, WNT1, EIF4EBP1, WNT7A, XRCC3, ZIC1, CNBP, TRIM26, EPHA2, EGR2, NTT, EDNRB, LPAR1, EPHB3, EPS15, ACP3, TSG101, TFAP2A, TFE3, FCER2, FBN1, FASN, TGFBR1, TGFBR2, EZH1, EWSR1, TK1, TLR1, TLR2, TLR3, TM7SF2, ETV4, ETS1, TP53BP1, TPI1, ERCC5, ERBB4, ERBB3, TPT1, CRISP2, HSP90B1, CCT3, SLC7A5, CND, TP53I3, TNFRSF10B, TNFRSF10A, E2F4, IQGAP1, SOCS2, PROM1, KSR1, CDK5R1, PER2, CCNA1, BTRC, MAP3K14, SOCS3, USP6, OSMR, BUB3, AURKB, PTTG1, CRLF1, DHRS3, STK17B, KLF4, COX5A, ATG5, ADAMTS1, PTGES, SCEL, CDC23, NAA10, BECN1, TKTL1, BAP1, CDC45, FZD1, FZD4, PLA2G6, TAGLN2, RECK, CAVIN2, CUL3, TYMP, MAP4K3, PARG, APOL1, LMO4, LGR5, CDC14B, KHSRP, PSMG1, ECE1, EIF4EBP3, E2F6, SOCS1, PDE5A, IRS2, CDR3
-
Thyroid Cancer
Mayo_clinic
Radiation therapy treatments to the head and neck increase the risk of thyroid cancer. Certain inherited genetic syndromes. Genetic syndromes that increase the risk of thyroid cancer include familial medullary thyroid cancer, multiple endocrine neoplasia, Cowden syndrome and familial adenomatous polyposis.RET, BRAF, KRAS, PRKAR1A, NRAS, FOXE1, LGALS3, CTNNB1, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PPARG, PAX8, CCDC6, TAS2R38, VEGFA, PTCH1, TSHR, TPO, PTEN, TP53, TG, TERT, TPCN1, SLC5A5, LOC110806263, EGFR, CXCL8, HT, F9, AKT1, CDKN2A, CD274, MTOR, MAPK1, XRCC1, STAT3, GSTT1, SGSM3, MAP2K7, XRCC3, PROM1, EPHB2, MIR146B, CALCA, NKX2-1, TGFB1, RASSF1, NTRK1, IGF1, NOTCH1, FGFR2, MMP2, GSTM1, GSTP1, ALK, THRB, KRT19, NCOA4, MTHFR, MUC1, TNFSF10, ERBB2, HRAS, SMUG1, CYP1A1, LOX, RNH1, NFE2L2, S100A4, RELA, GDF1, SLC26A4, MAP2K1, RAF1, EGF, EZH2, HIF1A, HABP2, IL10, CYP19A1, TNF, SYT1, RUNX2, GORASP1, SHC3, WNK1, ATM, CCND1, BDNF, PPP1R13L, PTTG1IP, YAP1, TMPRSS4, PTCSC3, ZEB1, THBS1, H3P23, IL6, IGF1R, ARHGAP24, AFAP1-AS1, IFI27, IDH2, FN1, IDH1, ICAM1, FOLH1, TMED7-TICAM2, UCA1, FUCA1, PRIMA1, SNHG15, FOXA1, TCIM, HMGA1, TICAM2, GSTK1, MALAT1, MIR126, MIR21, GABPA, MIR221, SLCO6A1, SLC2A1, KDR, IQGAP1, FHIT, RARB, RAP1GAP, TP73, PTH, TPM3, TWIST1, PSMD9, VEGFC, HMGA2, RASAL1, BECN1, PKM, SLIT2, ZNF395, ZNRD2, TXNRD2, DCTN6, PAK1, CHEK2, DICER1, MYC, PATZ1, SOSTDC1, HPGDS, MET, SST, LCN2, TMED7, H3P10, ECM1, CCR6, PARP1, APC, ESR1, CD44, ALB, FGF2, SEC62, THRA, SPHK1, TFF3, TAF1, STRN, STK11, STC1, CD40, TP53BP1, SPP1, TSC2, CCND2, TXN, UBE2N, UVRAG, VCP, VDR, CAV1, VIM, WT1, CXCR4, BAP1, CDK2, CDK5, SOD3, SERPINB5, MBL2, MLH1, MMP1, MMP9, COX2, CYP24A1, NTRK3, CYP3A4, PCNA, PDCD1, PDGFRA, POU5F1, CDK4, MAP3K8, PTGS2, PTPN11, NECTIN1, RAD51, RPE65, SAI1, CCL2, SDHB, SDHD, TP63, PPM1D, PTTG1, INPP4B, MIR27A, SNHG7, CCDC80, KLB, DACT2, MIR146A, MIR148A, MIR150, MIR19A, MIR20A, MIR204, MIR205, MIR338, FGFR4, MIR375, MIR497, MIR524, POU5F1P3, POU5F1P4, GRK2, MIR625, HOTAIR, VTRNA2-1, KLLN, MTCO2P12, MUL1, NDRG2, SMURF1, GOPC, TACSTD2, IL32, KL, GDF15, BAG3, NR1D1, HDAC9, AKT3, TXNIP, PDPN, WDR3, CKAP4, BCL2, ZHX2, MMRN1, SIRT3, SIRT1, CBX7, LINC00312, ZMYND10, KRT20, FBLIM1, FAS, SMAD4, NAT2, PTK2B, DUSP6, LAMB3, DPP4, RCAN1, ESR2, HSP90AA1, TNC, H2AX, EIF4E, HDAC2, ETV6, GSTM2, EIF1AX, IGF2, GOT2, GPER1, IL4, FOS, CXCR1, FOXO3, NRG1, LEP, SASH1, PELI1, AICDA, CXCL1, INTS2, XPO5, SRGAP1, MIB1, SLURP1, SEMA6A, KLHL14, NCOA5, ANKRD36B, HAMP, CXCL16, OGFR, FABP4, UBE2C, ACE2, MREG, ZNF654, FBXW7, LGR4, RNF139, SAGE1, IL17RB, HIF1AN, WDR11, MR1, TMEM184C, PAG1, USE1, AR, AQP4, BCAT2, PCBP4, IL21, LGR6, ATG16L1, ULBP2, RHBDF2, VTCN1, TCEAL4, WLS, FHOD3, GADD45G, PPP1R2C, LIMD2, ALX4, SPRY4, GPX3, RASSF5, FAM107B, FSD1L, MRO, ING5, TCTN1, GRB14, BHLHE41, FSD1, SDS, GAS5, C14orf93, TINAGL1, SOX17, INF2, HMGB1, KLK3, CREB3L2, MARCKSL1, APRT, FRTS1, UBE2Z, GGCT, MAPKAP1, PIWIL2, RASIP1, TUG1, LAT, LRIG1, POLDIP2, NOC2L, FGF21, EHF, FOXD3, NOX1, TSPAN13, BAAT, DKK3, SNX5, IL37, ARSA, ARR3, RGCC, HIPK2, ATF1, POT1, RNF19A, DKK1, SIRT4, HGF, NEMP1, ANGPTL2, HDAC1, LPAR3, ATP5F1E, ATIC, SNHG1, PPP1R15A, TRIM29, TBC1D9, PRDX5, METTL7A, ABI3BP, PYCARD, RHOA, TFCP2L1, CXXC5, IL23A, GINS2, WWOX, ARG1, ARAF, FXYD5, CPSF2, ERRFI1, MIEF1, GYPA, SLC35F2, ETFA, TRIM44, GATAD2A, SYTL2, SIRT7, REV1, HLA-G, SLCO2B1, COPZ1, TDGF1P6, OBP2A, SPANXA1, ADGRE2, STOML2, DUOX2, NOX4, DELEC1, IL22, F11R, FOXP3, ARG2, NDUFA13, SDF4, DNMT1, RTN4IP1, PROK1, NR2F1-AS1, VN1R17P, ZFAS1, GPR166P, MIR326, MIR330, MIR335, GAPDH, MIR340, MIR369, MIR370, MIR361, GAP43, MIR383, DUXAP9, MIR429, MIR451A, AFM, NRARP, MIR7-3, FYN, MIR7-2, MIR211, MIR212, MIR214, MIR217, MIR218-1, MIR218-2, AHR, MIR222, MIR25, MIR26B, GAS6, MIR296, MIR299, MIR30A, MIR30D, MIR34A, MIR7-1, FAP, MIR510, CARD11, MANCR, MIR4319, PROX1-AS1, CCAT1, MICA, SPRY4-IT1, FALEC, OPCML-IT1, BANCR, LINC00210, FOXD3-AS1, PANDAR, DARS-AS1, PTCSC2, LINC01410, FOXM1, ACVR1B, ACTB, PARP4, MIR1225, FGF3, FLT3, RASSF10, FRA16D, PARP4P2, LOC646736, FOSB, CXADRP1, MIR539, MIR592, MIR622, MIR639, MIR650, SSX2B, UNC5B-AS1, FGF13, FGFR1, MIR873, TNRC6C-AS1, MIR206, GDNF, GEM, ANXA5, GPR151, ANXA1, MUC15, ANPEP, SPC24, HORMAD2, GPD2, CBLL2, DEUP1, SLC5A8, OXER1, ZCCHC12, ZNF367, METTL7B, FLCN, ANO5, DIPK2A, PPARGC1B, LRRK2, GH1, MRGPRX4, ANXA6, FOXD2-AS1, HCP5, MASTL, GPX1, MIR22HG, MTDH, DNER, PKHD1L1, PRDM6, PRRT2, CYTOR, LINC00313, LMTK3, VASN, NIBAN1, MRGPRX3, USF3, UNC5B, GPRC6A, CTAG1A, MIR132, MIR137, MIR139, MIR143, MIR145, GNAS, GPC3, GJB3, MIR154, MIR15A, MIR17, MIR18A, MIR182, MIR184, MIR196A2, MIR197, GHRH, ALDH1A1, MIR125A, MIR106B, ZNF677, CASC2, MRGPRX1, NEAT1, AMH, ATG9B, CLEC4D, RSPO2, AMBP, MIR106A, BMP8A, ALOX15, ALOX12, IYD, LINC01194, MIRLET7C, MIR100, ERCC5, HMOX1, LILRB1, PTMAP4, PTPN2, JUND, PTPRF, PTPRJ, JUNB, JUN, RAD52, ITGAM, RAP1B, CLU, RCC1, RASGRF1, RBBP4, OPN1LW, CETN1, CEACAM5, RGS4, PTMS, PTMA, PRKAB1, CD82, PRKCA, PRKCB, PRKCQ, PRKDC, CRYGD, MAPK3, MAPK8, CRK, CREBBP, EIF2AK2, PROX1, DUSP4, COL11A2, COL1A1, CNTN1, PTGDS, CCR7, RLN2, RNASE1, RNASE3, CDKN2B, ILK, SLC6A9, SLC7A1, SLC16A2, SLIT3, SNAI2, HLTF, SNAI1, IL18, SORD, SOX9, SOX11, SPG7, IL17A, SRC, SRP72, IL13RA2, IDO1, SLC1A3, SKP2, S100B, ABCE1, ROS1, ITGA3, RPL36A, S100A1, SORT1, CDKN1B, IRS1, SHH, SCN10A, IRF5, CCL15, CCL20, INSR, CXCL10, SGK1, CRYZ, PRKAA2, SSX2, MKI67, DNMT3A, NQO1, MMP7, LASP1, MMP11, MMP14, MRC1, ABCC1, CITED1, MST1, MT1A, MT1G, MT1M, CYB5R3, MYO1F, DECR1, MUTYH, LDHA, MGMT, PRKAA1, KITLG, LIMK1, SARDH, LOXL2, LPA, LRP4, BCAM, LGALS1, EPCAM, SMAD3, LETM1, MC1R, MCL1, MCM7, MDM2, MEN1, DIAPH1, MFAP1, MVD, DDT, MYCN, MYH9, CXADR, PECAM1, PEG3, PGR, ABCB1, SERPINA1, KIR3DL1, CTLA4, CCN2, CTAG1B, CSF1R, PITX2, CSF1, KIR2DS5, POU5F1B, MAPK14, PPP6C, KISS1, KIT, KIF22, NOVA1, NCAM1, DDIT3, NFIL3, NFKB1, NNMT, AKR1C2, NOTCH3, ACE, SERPINA5, NRCAM, L1CAM, TNFRSF11B, PA2G4, PAEP, PRKN, PCBP1, SSTR3, CDH6, HPSE, SPAG9, SLC16A4, SLC16A3, CCNE2, HSPA9, PDLIM7, S1PR2, MMP20, TRIP13, BRCA2, UBE4A, HSPA5, GRAP2, ABCG2, GSTO1, MAP4K4, ELK1, HPT, CLDN1, SOCS3, BUB1B, AKAP4, ID3, AGPS, BHLHE40, MADD, ID1, PTCH2, NCOA1, BUB1, PEA15, CDC23, MBTPS1, BTG1, RAB11A, ELF3, KLF5, BSG, KSR1, HOXC10, CLOCK, HNRNPF, RASSF2, SPRY2, CITED2, BASP1, BMP4, HMGN4, SEC23B, CIB1, BMP1, NPC2, BLM, AHSA1, ERCC2, LIG4, POSTN, IGF2BP3, BID, CEACAM1, CTDSPL, TRIM13, PSME3, NR1I3, EIF4A3, TOMM20, KEAP1, PJA2, ZBTB5, FGF19, MED12, RBX1, PDZK1IP1, REC8, HNRNPD, HDAC6, ABCB6, HUWE1, NAMPT, AKAP9, TTF2, CAVIN2, DUSP5, TIAM1, TIMP3, TIMP4, CD40LG, IL1B, ABO, IL1A, IGSF1, CD34, MS4A1, CD3D, TRPC1, TSC1, IGFBP7, TSG101, CCNG2, TTF1, IGFBP4, TIMP1, IL1R1, RECK, IL2, STK3, STK4, CXCR2, CDH5, TBX1, TAZ, TBX15, TCF3, CDH1, TCP1, TEK, TEP1, ADGRE5, TFAP2B, CD74, CD68, CD63, IGFBP1, TXNRD1, UCHL1, UPP1, YY1, ZIC1, ZNF20, BTG2, CASP8, IL1R2, AIMP2, CAMP, DDR1, SHOC2, TSPO, IFT88, SLC7A5, TKTL1, ELAVL1, FZD4, FZD8, YES1, XRCC5, XRCC4, IFNA13, USF1, USF2, EDN1, EFNB2, VDAC1, IFNG, RUNX3, VHL, CASR, EZR, IFNA1, WIPF1, WRN, XIST, CAT, XRCC2, FH
-
Breast Cancer
Wikipedia
Cancer that originates in the mammary gland Breast cancer Mammograms showing a normal breast (left) and a breast with cancer (right) Specialty Oncology Symptoms A lump in a breast, a change in breast shape, dimpling of the skin, fluid from the nipple , a newly inverted nipple, a red scaly patch of skin on the breast [1] Risk factors Being female, obesity , lack of exercise, alcohol, hormone replacement therapy during menopause , ionizing radiation , early age at first menstruation , having children late in life or not at all, older age, prior breast cancer, family history of breast cancer, Klinefelter syndrome [1] [2] [3] Diagnostic method Tissue biopsy [1] Mammography Treatment Surgery , radiation therapy , chemotherapy , hormonal therapy , targeted therapy [1] Prognosis Five-year survival rate ≈85% (US, UK) [4] [5] Frequency 2.1 million affected as of 2018 [6] Deaths 627,000 (2018) [6] Breast cancer is cancer that develops from breast tissue. [7] Signs of breast cancer may include a lump in the breast, a change in breast shape, dimpling of the skin, fluid coming from the nipple , a newly-inverted nipple, or a red or scaly patch of skin. [1] In those with distant spread of the disease , there may be bone pain , swollen lymph nodes , shortness of breath , or yellow skin . [8] Risk factors for developing breast cancer include being female, obesity , a lack of physical exercise, alcoholism , hormone replacement therapy during menopause , ionizing radiation , an early age at first menstruation , having children late in life or not at all, older age, having a prior history of breast cancer, and a family history of breast cancer. [1] [2] About 5–10% of cases are the result of a genetic predisposition inherited from a person's parents, [1] including BRCA1 and BRCA2 among others. [1] Breast cancer most commonly develops in cells from the lining of milk ducts and the lobules that supply these ducts with milk. [1] Cancers developing from the ducts are known as ductal carcinomas , while those developing from lobules are known as lobular carcinomas . [1] There are more than 18 other sub-types of breast cancer. [2] Some, such as ductal carcinoma in situ , develop from pre-invasive lesions . [2] The diagnosis of breast cancer is confirmed by taking a biopsy of the concerning tissue. [1] Once the diagnosis is made, further tests are done to determine if the cancer has spread beyond the breast and which treatments are most likely to be effective. [1] The balance of benefits versus harms of breast cancer screening is controversial. ... Indications of breast cancer other than a lump may include thickening different from the other breast tissue, one breast becoming larger or lower, a nipple changing position or shape or becoming inverted, skin puckering or dimpling, a rash on or around a nipple, discharge from nipple/s, constant pain in part of the breast or armpit and swelling beneath the armpit or around the collarbone. [20] Pain (" mastodynia ") is an unreliable tool in determining the presence or absence of breast cancer, but may be indicative of other breast health issues. [17] [18] [21] Another symptom complex of breast cancer is Paget's disease of the breast . This syndrome presents as skin changes resembling eczema; such as redness, discoloration or mild flaking of the nipple skin. ... The risk is not negated by regular exercise, though it is lowered. [50] There is an association between use of hormonal birth control and the development of premenopausal breast cancer, [32] [51] but whether birth control pills actually cause premenopausal breast cancer is a matter of debate. [52] If there is indeed a link, the absolute effect is small. [52] [53] Additionally, it is not clear if the association exists with newer hormonal birth controls. [53] In those with mutations in the breast cancer susceptibility genes BRCA1 or BRCA2 , or who have a family history of breast cancer, use of modern oral contraceptives does not appear to affect the risk of breast cancer. [54] [55] The association between breast feeding and breast cancer has not been clearly determined; some studies have found support for an association while others have not. [56] In the 1980s, the abortion–breast cancer hypothesis posited that induced abortion increased the risk of developing breast cancer. [57] This hypothesis was the subject of extensive scientific inquiry, which concluded that neither miscarriages nor abortions are associated with a heightened risk for breast cancer. [58] Other risk factors include radiation [59] and circadian disruptions related to shift-work [60] and routine late-night eating. [61] A number of chemicals have also been linked, including polychlorinated biphenyls , polycyclic aromatic hydrocarbons , and organic solvents [62] Although the radiation from mammography is a low dose, it is estimated that yearly screening from 40 to 80 years of age will cause approximately 225 cases of fatal breast cancer per million women screened. [63] Genetics [ edit ] Genetics is believed to be the primary cause of 5–10% of all cases. [64] Women whose mother was diagnosed before 50 have an increased risk of 1.7 and those whose mother was diagnosed at age 50 or after has an increased risk of 1.4. [65] In those with zero, one or two affected relatives, the risk of breast cancer before the age of 80 is 7.8%, 13.3%, and 21.1% with a subsequent mortality from the disease of 2.3%, 4.2%, and 7.6% respectively. [66] In those with a first degree relative with the disease the risk of breast cancer between the age of 40 and 50 is double that of the general population. [67] In less than 5% of cases, genetics plays a more significant role by causing a hereditary breast–ovarian cancer syndrome . [68] This includes those who carry the BRCA1 and BRCA2 gene mutation . [68] These mutations account for up to 90% of the total genetic influence with a risk of breast cancer of 60–80% in those affected. [64] Other significant mutations include p53 ( Li–Fraumeni syndrome ), PTEN ( Cowden syndrome ), and STK11 ( Peutz–Jeghers syndrome ), CHEK2 , ATM , BRIP1 , and PALB2 . [64] In 2012, researchers said that there are four genetically distinct types of the breast cancer and that in each type, hallmark genetic changes lead to many cancers. [69] Other genetic predispositions include the density of the breast tissue and hormonal levels. ... The familial tendency to develop these cancers is called hereditary breast–ovarian cancer syndrome . The best known of these, the BRCA mutations , confer a lifetime risk of breast cancer of between 60 and 85 percent and a lifetime risk of ovarian cancer of between 15 and 40 percent. ... The inherited mutation in BRCA1 or BRCA2 genes can interfere with repair of DNA cross links and DNA double strand breaks (known functions of the encoded protein). [86] These carcinogens cause DNA damage such as DNA cross links and double strand breaks that often require repairs by pathways containing BRCA1 and BRCA2. [87] [88] However, mutations in BRCA genes account for only 2 to 3 percent of all breast cancers. [89] Levin et al. say that cancer may not be inevitable for all carriers of BRCA1 and BRCA2 mutations. [90] About half of hereditary breast–ovarian cancer syndromes involve unknown genes. Furthermore, certain latent viruses, may decrease the expression of the BRCA1 gene and increase the risk of breast tumors. [91] GATA-3 directly controls the expression of estrogen receptor (ER) and other genes associated with epithelial differentiation, and the loss of GATA-3 leads to loss of differentiation and poor prognosis due to cancer cell invasion and metastasis. [92] Diagnosis [ edit ] Most types of breast cancer are easy to diagnose by microscopic analysis of a sample - or biopsy - of the affected area of the breast.BRCA1, BARD1, PIK3CA, BRCA2, ESR1, TP53, CDH1, CAV1, PTEN, ERBB2, CHEK2, BRIP1, PALB2, ATM, EP300, FGFR2, AKT1, KRAS, RAD54L, PARP1, ROR1, STAT1, FOXA1, ESR2, NOS2, CASP8, AKT2, FGFR1, NOTCH1, HRAS, RB1, CDKN1B, FGF3, NCOA3, FBXW7, PPM1D, FN1, RAD51, IGF1, MMP1, MDM2, NQO1, PHB, PTHLH, AR, BAP1, GATA3, TBX3, FLT1, NOTCH2, GPNMB, RB1CC1, FGF4, IFNB1, PLA2G4A, NCOR1, PDPK1, ZNF366, MYH9, ADAR, SREBF2, ICAM5, APC2, GRIK2, STARD8, NRCAM, TOX3, MSH6, NBN, ERBB4, ABCC1, RAD51B, MDM4, FTO, ZNF365, COL7A1, ZNF432, NOP9, ARID1B, LSP1, CDKN2A, HOXB13, HADHB, LGR6, STXBP4, ZMIZ1, OLA1, NR2F6, SORBS1, YBX1, GPX1, XRCC3, AURKA, XRCC2, NAT2, CXCR4, GRB7, RECQL, CXCL12, SNAI2, GJA1, PIN1, KLK10, STMN1, BCL2, CCND1, DKK1, RAF1, XBP1, CAT, BAX, PHGDH, PIK3CB, MIR29A, GPER1, BMP2, MIR206, BRAF, BCAR1, ABCG2, MIR222, MIR221, GLI1, MIR214, APOBEC3B, ADAM12, CCNE1, WWOX, BMP4, MTDH, MIR200C, MIR200B, ZEB1, LOXL2, LEPR, LEP, SNAI1, AGR2, SULT1A1, WT1, CYP1A1, IFNG, PTGS2, ETV4, EZH2, CXCL8, TYMS, F3, CYP24A1, CYP19A1, CYP17A1, PAEP, CYP3A4, IL6, CYP2D6, IL24, ZEB2, CYP1B1, THBS1, FASN, TGM2, JUN, IL10, ESRRA, SLC2A1, E2F1, UBE2C, EGFR, EGF, TOP2A, SLC5A5, EDNRA, TP53BP1, TP73, IDO1, ERBB3, TNF, ABCB6, EPHB4, PAK1, DNMT3B, DNMT3A, SERPINB2, DNMT1, CTNNB1, KEAP1, CST6, RAD51C, COMT, IGF1R, TFAP2A, SNCG, MIR10B, YAP1, MIR126, KRT5, TERT, FOXM1, FOS, MIR141, MIR145, MIR146A, SERPINB5, CHEK1, MTOR, SFRP1, VIM, VEGFC, IL1B, TFRC, SLC39A6, FOXP3, MALAT1, CSF3, CSF2, CSF1R, CSF1, SIRT1, KDR, NISCH, PGR, IGFBP7, IGFBP5, ABCB1, CLDN4, VDR, BCAR4, NDRG1, BAG1, MIR205, NOTCH3, ABL1, H2AX, ADAM10, APRT, MST1, CLDN1, MMP9, NCOA1, PRKAA1, SETD2, AREG, TNFSF10, PRKAA2, STAT5A, INPP4B, GSTP1, HPSE, PDCD4, JAG1, SPP1, MAP3K1, ARID1A, RARB, MMP14, STAT3, NOS3, MTR, MKI67, MMP3, MMP2, HSP90AA1, AHR, NRG1, HRG, HIC1, HIF1A, MRE11, RARA, SOD2, SRC, MTHFR, MAP2K7, NOTCH4, NHS, BCAR3, APC, GSN, HMOX1, ETS2, TUBB3, PER2, KCNIP3, CUX1, NF1, ADRA1A, WNK1, PPARGC1B, RELA, TEK, DHFR, KIT, MIR342, SLC2A5, ANKRD30A, IKBKB, CADM1, MIF, MIR152, HMMR, NRIP1, SLCO1B1, SFRP2, KRT8, HES1, APOBEC3A, EMSY, RPS6KA3, MIR429, EPOR, DDX3X, FLNA, GPI, LEF1, TXN, RNF115, PIM1, CD40, FABP4, FST, ATF2, LAMTOR5, ALK, MIR127, RASAL2, CDH5, PLXNB1, PER3, SLC22A18, KDM3A, CXCR5, MIR301A, MAP2K4, PTGS1, RBM3, CPT1A, DLL4, KCNH1, CD74, PRC1, MIR489, CFL1, NET1, DLEC1, NCOA2, BIRC5, H19, ENO1, SGK3, KRT14, NF2, TOP1, MME, KRT18, MIR132, PER1, TIMELESS, C1QBP, RAPH1, FHL2, TFPI2, NR2F1, ATP2A3, MIR10A, CCL20, HP, MFGE8, NEDD4, PFKFB4, JMJD6, FOLR2, SULF2, WNT10B, FGF10, GPX4, SLC16A3, STC2, DDIT3, BCL2A1, HOXB9, RPS6KB2, OCLN, ABRAXAS1, EIF4A2, LHCGR, SHMT1, PRDX5, FLNB, RPS6, GAB1, PBRM1, LDHB, GZMB, MYOD1, BCHE, KMT2D, CDKN2C, CLSPN, FOXQ1, PDLIM7, GPX2, ARRDC3, MSI1, CLCA2, CSNK1D, ACHE, DEK, ECT2, PLD2, ADAMTS1, NORAD, DPYD, ELK3, H6PD, PTPRC, THEMIS2, FKBPL, PLS3, TACC2, AKAP9, CENPF, CNR2, NAA25, SERPINE2, SLC35A2, GPC1, ROBO1, HDAC4, MAL, ABCA4, BIRC2, BMPR2, XDH, RRAD, CBFB, CXCL9, CLIP1, FABP7, ARFGEF2, TNIP1, HDLBP, TENM1, MED28, SFRP5, EFNA1, DSC3, BCL11A, NFKBIA, TMPRSS6, SLCO2B1, SRARP, RARG, EXO1, APCS, NQO2, ACVR1, ERCC6, MAOA, ANGPTL4, FBL, HDAC7, ADAM33, TRIM29, CDC27, HHEX, MECOM, AKAP12, EPB41L3, LPAR1, OCA2, GABRP, CDA, CUL5, IRF8, LIMD2, DES, KLK15, ARF1, SRSF1, SLC2A2, NSD2, RGS2, GSDMB, EREG, ENPEP, PPM1E, FOXP4, LLGL2, EFEMP1, MTMR3, EIF2S2, NUP214, AMFR, MAGEA4, PCDH8, FGD5, MIR101-2, DDX10, BTK, REPS2, ARAP3, ZNF404, TP53BP2, PLCB1, EHMT1, MEIS1, HSD11B1, MAP3K13, FBXO8, NCOA6, ATG10, CASP7, BGN, CHD5, CIC, TCL1B, ACCS, FAAH, CTU1, NUP98, ARHGDIA, TCP1, MICAL1, FUS, PDCD6, H4C1, EDNRB, SEMA3A, MMP10, RPS4X, AFP, DLL1, MIR24-2, ABCB10, UTS2R, MFAP5, CDH2, IFNA2, DIP2C, DAP3, GEN1, ZFYVE26, UMPS, KCNA5, CLPTM1L, CFP, SUV39H2, SLC2A10, SLC10A6, BCORL1, DMD, BTN3A2, HNRNPK, HNRNPL, PTPN14, HOXA4, GPC2, HEY2, SUSD3, PTPRD, ACADM, DVL3, TICAM1, ZNF569, MAGEC2, B3GALNT2, SLC6A3, MT3, NDUFS3, TEKT4, ERCC3, DIO3, P3H2, NIPBL, PRRT1, ZNF436, ARHGAP11A, HLCS, FLACC1, JAG2, AHSA2P, SIM1, PDZK1, ZC3H11A, SLAMF1, SRGAP3, SPATA18, G3BP2, MACF1, NSMCE2, AKAP8, MED12, RIC8A, ADHFE1, PDGFA, DEPP1, SLC28A1, RMND1, HEYL, IBSP, RSPO3, SLC6A5, DOP1A, AQP8, PRODH, CFL2, MAGEA1, HELQ, RPRD1A, POU2F1, RXRB, RPS9, WARS1, LBX1, BMP1, GALNT16, COTL1, MRPS23, ASL, DHX32, ZNF668, SCP2, TP53I3, HSPA1B, CCNH, PPA1, ELP3, TRERF1, H2BC4, RCCD1, SPAG6, TBXAS1, LIPE, UBD, SIK3, KAT6B, VWF, MNX1-AS1, PRPS1, TREM1, MIR31HG, MAGED2, MCF2L2, CXCL3, RAC2, SYNJ2, APPL1, VEGFB, CRHR1, LRRC3B, MANEA, PRPF4B, GRIK3, CEP85L, COL11A1, EPG5, NMBR, TAS2R13, GKN1, HERC2, SMC6, INPP5K, MACIR, C16orf58, HEY1, RUFY1, SMARCD1, EIF5, CELA1, SPTLC1, SLC9A2, FNDC3B, CXCL2, SYT3, TLL1, NLE1, SMARCA4, GRIN2D, GDF10, SPAST, ZNF541, EVL, HOXA3, GRM6, TSPAN4, STX5, NECTIN2, PLEKHA8, GOLGB1, ANKRD29, NOA1, FARP1, SURF1, RAP1GAP, RFC4, MICALL1, LZTS2, B4GAT1, NPBWR1, L3MBTL3, STX12, DYNC2H1, MFSD9, TMEM25, UNC45B, SLC8A3, HOOK3, NFYC, ABHD12B, POP1, GNAI2, TMTC4, WDCP, SMOX, H2BC12, KBTBD8, SIX4, RANBP1, ARHGEF38, ZBTB3, LMF2, STPG1, PKDREJ, OSBPL11, SORCS1, HGF, GNB1L, THBS3, FCRL5, TESK1, UGT1A9, TCF7L1, IGBP1, FAM83F, ADAT3, HOXD11, DDX59, COG3, TECTA, CPSF3, EIF3M, ZNF281, FCRL3, PCDH20, PIGS, TMEM125, TSC22D4, MRPL9, CFHR5, FAM189B, DNAJC24, HK3, EFCAB13, SORL1, FAM210A, SCGB3A2, NLRP8, WDR88, FKTN, KLHDC7A, AGAP2, OR12D3, C1orf87, PPHLN1, ZNF318, INHBE, H1-2, GTF2A1, SPTAN1, CD109, ARFRP1, ALKBH8, BOC, MPLKIP, PHF7, RASL10B, RALYL, LONRF3, SCARF2, ZNF644, SEMA5B, NLRC5, FASTKD3, PUS1, SPOCD1, TMEM39A, TLE3, HNRNPR, SLC25A51, FSCB, TFAP2D, UBLCP1, FAM131A, TXNDC15, METTL6, RTP1, TRMT10A, VPS72, RASGRF2, OBSCN, LDHAL6B, G6PC, GUCY2F, STAT4, GABRA4, PADI3, GUCY1A2, ATP6AP1L, TMED1, DNAJC21, SP110, SLC22A9, OR1N1, DTX3L, PHKB, CLCN1, CLIC1, ZNF532, KPNA5, GALNT5, MAGEE1, CNGA2, ZSWIM5, PPP1R12B, OXSM, ZFP64, SHROOM2, ZNF646, ABCB8, SMARCAL1, PDZD4, SPATA21, KANSL1-AS1, GPR45, CDS1, KIAA1324, PIK3R4, LRRC7, LAMA2, NUP133, WNT1, KTN1, MET, PPM1F, TRMT2A, CHRND, ARAF, YY2, HAPLN4, MRPL13, KRTAP10-8, SETBP1, ITSN2, OSTC, ZNF277, ARHGEF4, CRX, SIPA1L1, COL19A1, CDH20, CRYAA, KCNJ15, C5orf34, PRRC2A, MROH7, LINC00671, KALRN, C4orf50, GFRAL, WFDC1, MRPS28, NUDT2, ACAP1, ARTN, PPP1R3A, SLC7A7, PLEKHD1, APOC4, MAP3K6, MIA2, WBP4, N4BP2, INA, VPS39, EOMES, MYH1, FEM1C, SLC17A6, MIR345, IL1RAPL2, CCT5, PURG, SCNN1B, MOSPD1, MAN2C1, TBC1D9B, LRRC37A2, PANX2, PLAU, MCOLN1, ARFGAP3, UBR4, CMC2, ZDHHC4, KLHDC10, CUBN, MRPS22, OTOF, ARHGAP29, C1QB, AKAP6, CNNM4, MYC, PLOD1, SPEN, KDM1A, ZNHIT2, FXR1, PCDHB15, RRP9, LGALS2, LRBA, LRRFIP1, TOR1AIP1, POLH, AVPR2, GGA1, POLR2F, PRICKLE3, ATP7B, NID2, ATP6V0B, CDH10, COLGALT2, DYSF, PRRG1, PCDHGB6, PABPC1, RIBC2, TAFA4, MAGOHB, RIF1, SYNE1, FRMPD1, ZNF22, LLGL1, ZNF75A, DAZAP1, PNLIPRP1, MED14, CD2, TRAPPC8, GGA3, KCNJ1, RHD, SH2D3A, TRPC4, DNASE1L3, NME8, BEND7, PXDC1, DPAGT1, RNF182, HEPACAM2, KIF6, EHBP1, RPL31, ADGRF4, NSUN6, SLC39A12, DNAH9, ACTA2, PGBD3, MAP7D2, MRPL19, RGL1, ALS2CL, CATSPERE, ITGA9, HEATR9, TTC3, HSPA14, ACY1, TANK, SNX32, RPLP2, NDUFA3, THOC5, ZNF438, ACO2, ZNF25, EEF2, SSNA1, GPR180, ABCA3, USP54, EEF1B2, RFX2, SYNE2, KRT76, MAMDC4, VPS13B, TNS1, EGR1, RPL23A, ASTN2, FAM217B, TP63, DAW1, ANKEF1, TPM4, ANAPC5, XIRP1, TPTE, GIMAP1, ADAMTS19, NIP7, DTX3, PACC1, UQCC1, NDUFA8, NUDT17, HOOK1, BOD1L1, RCE1, CNTN6, MRPS7, ALDOA, CLUL1, KCNT2, TMEM161A, TREML1, TRIML1, ELP1, KRTAP21-1, ZNF546, RPS7, NDUFA2, DGKE, NLRP14, STRBP, UPK1B, DDX18, CUTC, ZNF667-AS1, TAX1BP1, CTU2, AP1M1, RPS8, ATP8B1, AFF1, MTMR8, KIF16B, ANK1, WDR53, CST4, CTIF, ANK2, NLRP9, TRAPPC12, TRMT11, DGKG, JAKMIP3, PDE2A, CYB5R4, OTOGL, EXOC3L1, CAVIN1, ZCCHC14, SKIV2L, ANKRD34A, APOL1, GOLIM4, DBN1, CLASRP, RNF149, EIF6, LRRC37A, KRTAP20-1, H2AC21, DDO, HSD17B8, SBNO1, UTP20, THRB, ETV6, ESPL1, TCF7L2, ADSL, CASC16, MSH2, MRTFA, MLH1, MUTYH, SLC19A1, PVT1, PRSS46P, SLC4A7, TGFBR2, RET, CCDC170, PMS2, TET2, COL1A1, SIAH2, ITPR1, ARHGEF5, CASC9, RNF146, TAB2, BABAM1, ANKLE1, EBF1, CDKN2B-AS1, FANCI, PEX14, HSPG2, RALY, PDE4D, POU5F1B, DCLRE1B, CCDC88C, ELL, ADAM29, CASC8, FBXO11, SLC22A3, SOAT1, BRCA3, SLN, SMUG1, IL4, ZNF410, MMP13, MMP11, TBC1D9, JAK2, MMP8, MMP7, HSD17B1, IL2, NCL, NLRP2, CCN1, JUNB, JUND, COPS5, SHBG, MRPL34, IL1A, TRPM7, MUC1, CD82, NEU1, SIX1, IL17RB, IL17A, REN, TNIP3, HEATR6, MT1E, IL15, PGRMC1, RPE65, SOX9, IL18, RPS6KB1, SLC9A1, BRD4, IRS1, SOX17, CXCL10, ITGA6, INSR, COX2, GAS5, IL11, CXCR2, SKP2, MPO, MEG3, RASSF1, SLC16A1, SOX2, ITGB3, NEK2, LILRB1, HPGD, ITGB1, SOX4, MRC1, LAPTM4B, NME1, MSH3, TRNE, MTRR, IGFBP3, RRAS, ROS1, LPA, MAPT, RETN, S100A9, CCL2, S100A8, S100A7, S100A6, S100A4, LOX, MCPH1, CCL5, LNPEP, CCL18, VTCN1, HTC2, LMNA, MCAM, IFI27, BECN1, NFKB1, MGMT, SMAD3, UGT1A1, MUL1, MYBL2, EPCAM, SUB1, SLC12A9, SLC52A2, SMAD2, SMAD4, S100B, SATB1, TNC, ACKR3, SCD, MYB, ABHD8, MYD88, FSD1, CCL21, MCL1, LIG4, ANO8, SET, SEMA6A, PREX1, KRT19, MFAP1, DDA1, CIP2A, RRM1, IGF2, SLC52A1, KISS1, SCGB2A2, IGF2R, HSF1, ICAM1, RNASE3, BANP, IGFBP2, SELE, INTS2, LASP1, HSPA4, MYO9B, SIRT3, NGF, LIN28A, SDC1, CKAP4, LGALS3, NFE2L2, HSPB2, HSPB1, LGALS1, SHROOM4, LDHA, LCN2, PHB2, PPARGC1A, HSPA5, DICER1, NAT1, SP1, MIR29B1, MIR34A, MIR31, MIR30C2, CASP3, MIR30A, NPEPPS, CASR, RUNX2, RUNX1, PSMD9, MIR29B2, RUNX3, MIR27B, CMA1, CCK, MIR27A, ZNF217, PLAT, CCNB1, PSAT1, CCND2, PLAG1, MIR223, MIR22, NR1D1, CLOCK, PLG, BTG2, CALR, MIR93, MIR373, PPIG, KL, PML, MIR340, CD274, SLC9A3R1, ADIPOQ, BSG, PSG2, MIR335, KLF5, COX5A, BTF3P11, GRAP2, CAPN5, GSTO1, AIMP2, CA9, CA12, USP7, CALCB, CALCR, MIR96, PLK1, MIR210, MIR21, NCOR2, XPC, CDKN1A, PIK3R1, CDKN2B, PIK3CG, PIK3CD, CEACAM5, MIR182, MIR18A, CEACAM3, CEACAM7, MIR17, IKBKE, SERPINB6, MIR155, WNT5A, MIR148A, CHI3L1, CHKA, MIR143, MIR142, MIR140, CISH, CKS1B, MIR139, SPARC, PTH, CDK6, CD34, CDK4, CD36, MIR204, MIR203A, ZFP36, CD44, CD47, TRAF4, CD68, MIR200A, MIR20A, CD151, CDK1, MIR19B1, PKM, KLK4, CDC25A, MIR195, PITX2, CDC42, ERVW-1, XRCC4, PIP, MIR191, CDK2, XRCC1, BMP7, BMP6, FOSL1, PCAT1, ERVK-20, COMMD3-BMI1, LMO4, ADRB2, MAPK3, MAPK7, MAPK8, TMED7-TICAM2, AGER, AGT, AGTR1, NR1I2, ALB, ALCAM, ALDH1A1, CD24, PGAM1P5, SPHK1, AKR1B1, SQSTM1, HOTAIR, AMH, MBD2, MCAT, DIRC3, MAPK1, ERVK-18, PTPA, LINC-ROR, PRKCB, H3P10, H3P23, EIF3A, IRS2, PRKCA, DKK3, ABO, TNFRSF11A, ERVK-32, MTCO2P12, TNFRSF10B, TNFSF11, ACTB, PRKAB1, DLX2-DT, NRP1, LINC01488, LINC01527, LINC02143, LINC01416, PRKDC, ADH1C, HPGDS, ADM, CCR2, ANGPT1, BMI1, AXIN2, POU5F1, MIR499A, ATF3, MIR497, PROS1, LPAR2, PON1, TAM, ATR, HACD1, MIR20B, BACH1, RNU1-1, PTTG1, KLK6, BCL6, KLF4, HMGA2, MIR375, BDNF, CD163, CEACAM1, SLIT2, PSEN2, BLM, NEURL1, STS, ANGPT2, POU5F1P3, HSPB3, SGSM3, ANXA1, ANXA2, ANXA5, ANXA6, FOXP1, SOCS3, APEX1, XIAP, MCTS1, PELP1, APOE, PRL, KLK3, FAS, CLDN6, PRLR, DIRAS3, UCA1, PPARG, MTA1, EMBP1, POU5F1P4, PPARA, POLDIP2, CLU, CCR5, AGO2, NR5A2, MUC16, CAP1, FUT4, ODC1, CARM1, XRCC6, GABPA, GAPDH, MYBBP1A, CIB1, RAD51D, ZNRD2, PAK4, GH1, GHRH, TAZ, CXCL13, MAP3K7, ABCC11, GLS, ADAM17, NTRK1, TAC1, ARFGEF1, NT5E, PRRT2, PRMT5, OGG1, FOSB, EMID1, TGFBR1, FDXR, FEN1, FGF1, FGF2, SPDEF, FGF7, CDYL2, RAC1, FGFR4, TGFB1, TGFA, FHIT, MUCL1, OSM, TFF1, DLC1, GDE1, FOXC1, ANIB1, TFAP2C, FOXO1, FOXO3, TNFRSF11B, SYK, SFN, AHSA1, HDAC1, KHDRBS1, HFE, SSTR4, CXCR6, SST, HK2, HLA-A, CTCF, NECTIN4, DCTN6, HLA-G, COL18A1, HMGB1, NPY, SRCIN1, KRT20, HMGA1, HNF4G, CD276, NES, PPP1R2C, KDM5B, HOXA5, SEPTIN9, WASF3, ST14, ARHGAP24, CXCR3, PLVAP, PDLIM5, STK11, POSTN, GRB2, GRID1, GRID2, GRN, BRMS1L, NR3C1, NRAS, TLR9, CXCL1, GRPR, GSK3B, GSTM1, GSTM2, STAT6, STAT5B, GSTT1, NPAS2, USHBP1, FSD1L, BCL2L12, CHST9, HAS2, CCR7, PTK2B, FANCD2, CYP2C19, CTAA1, TMED7, CCN2, CTLA4, GMNN, CTSB, CTSD, CTSL, FMN1, CYP1A2, PTPN1, CYP2B6, PECAM1, PDCD1, TAS2R64P, PTPN11, CYP27B1, DACH1, PDK1, DAPK1, TWIST1, DCN, ACE, NEAT1, TTK, DHCR24, TICAM2, GSTK1, UGT2B7, PFN1, MIR125A, MIR122, GREB1, PTK2, MIR100, PTK6, LINC01194, COL11A2, VIP, EZR, LINC01376, COX8A, VEGFA, ADAMTS7P3, HDAC9, CLDN7, CPOX, LDLRAD1, CREB1, VASP, UVRAG, CRK, CRP, MAPK14, F11R, BRMS1, SCO2, DLX4, AQP4-AS1, SRA1, PWAR1, EPAS1, CFAP54, PRKN, CNTN3, EPHB2, TLR4, PXN, EPO, TLR1, ERCC1, ERCC2, ERCC4, ERCC5, ESRRB, SERPINE1, ETS1, TIMP3, TIMP2, TIMP1, F2R, F2RL1, SLCO6A1, SYNPR, CTTN, HSD17B7, CBLL2, RNF19A, TSG101, TRPS1, HDAC6, ATN1, PCNA, DUSP1, RAD50, E2F4, TYMP, C11orf65, NAMPT, CYTB, TNFRSF1B, NEK10, PAX9, EPHA2, TNNT3, ELF3, EIF4E, EIF4EBP1, TNFRSF1A, ELAVL2, MTNR1A, LPAR3, BBC3, PYCARD, RNU1-4, WWTR1, MYOF, PAWR, GDNF, PRKD1, CCAR2, CTNND1, IGFBP1, FERMT2, IL1RN, UGCG, CTBP1, CTAG1B, CXCR1, IL13, VCAN, SMURF2, VCAM1, GPR42, ING1, VIPR1, MIR137, MIR149, ITGA5, PTP4A3, ZNF350, CDKN3, MELK, IFNA13, IFNA1, GRHL2, HAVCR2, SULT1E1, TCHP, STC1, GLO1, SSTR2, STARD13, SCGB3A1, VEGFD, SOX10, NR1I3, NANOG, TLR2, EPHA3, TRIM28, TPT1, DUSP4, FSCN1, CYP4Z1, DDX5, CDKN1C, ILK, CDK7, FANCM, BRS3, BCS1L, PIWIL1, XPR1, MIR193B, CCNE2, MEN1, MIR320A, RRM2, CUL4A, PROM1, ADRA2B, CCN5, RHOBTB2, GDF15, ZNF469, CDH13, CCND3, PCGF2, MIR26B, CDC25C, KMT5A, FGF8, MIR183, P2RX5, KLK5, TGFBI, LMTK3, RECK, ANXA3, CKS1BP7, ESRRG, TFPI, FOLH1, MAP2K1, LINC01672, CEBPD, GDF2, TBX2, CCN6, SYT1, ORAI1, CDK10, LOC107987479, ABCC3, CDX2, TXNRD2, CTCFL, ARF6, TLR3, CRY2, CRYAB, MIR34B, MIR30C1, DDR1, CASP9, RASSF7, BNIP3, MIRLET7B, PRSS50, MIR29C, POLD1, ADIPOR1, AXL, USP9X, GAB2, UBE2I, PTCH1, MIR15A, TSC2, CTAG1A, ARNTL, LATS1, MED1, WRN, PAPPA, AKT3, WWC1, SMYD3, RBP1, RPS6KA1, TRIB3, MGP, S100A1, MCC, NDRG2, RAB40B, HOXB7, SAFB, HOXD13, WBP2, MRPS30, SOD1, RAB25, CYP4F3, PRMT1, MMRN1, MSMB, MTA3, IL2RA, MT1JP, FRTS1, RASA1, GORASP1, SLC20A1, PPP1R1B, MT1F, NEDD9, C17orf97, CAMP, LRP1, RFC1, CXCL5, CREBBP, ANO1, S100A14, LIFR, CERS2, CXCL14, CALM1, KDM6A, CCNA2, LIMK1, IRF1, PHLDA1, MIR150, GOLPH3, FSTL1, CEBPB, MIR185, PSIP1, KMT2C, CDK9, ABCC2, CDH3, MIR106B, TSPO, PKD1, GRM1, CDC20, YWHAZ, CD59, LGALS7, MIR17HG, TXNIP, BUB1, MICA, APP, MIR590, APOA1, KMT2A, RBM38, NR0B2, DYRK2, ALDH1A3, MSN, HDAC3, RPS6KA6, UHRF1, MT1A, MT1B, BHLHE40, ADH1B, MT1H, IQGAP1, ACACA, IL17B, SOCS1, MT1L, FASLG, LGALS7B, PRNP, RHOA, BST2, LTA, BIK, REST, INTS6, MIR423, MIR424, KDM4B, MAD2L1, SMAD7, BAD, ROCK1, MIR451A, ATP2B2, ARHGEF2, POU1F1, MBL2, POU4F2, MIR503, FZD4, ZNF423, LUM, EBI3, STARD3, ENG, PVR, MT2A, FLT4, ELF5, EIF4G1, TPD52, OXER1, DNM1L, NR2C2, HSP90B1, TRAF2, TNFRSF12A, GPRC6A, RAC3, ERP29, HAX1, TIMM8A, PART1, TM7SF2, DNER, USP17L2, PRAP1, SIRT6, POLQ, HNF4A, TET1, TFF3, FBLN1, FOXF1, PEBP1, FABP5, PRDX1, HOXD10, RBBP8, FLOT2, HMGCR, PIWIL4, AGFG1, OTUD4, PLAC1, DCTN4, CYP11A1, PTPN6, CYP3A5, CYP2E1, GADD45A, GHR, GLI2, GGCT, NFATC2, STING1, UCHL1, IL6ST, PFKFB3, DAPK2, GTF2H1, GRP, STIM1, NUP62, TMPRSS13, LMLN, ATG7, TCF4, PDGFRB, TXNRD1, MVP, ING4, G6PD, MOK, RIPK1, RXRA, CLDN2, TERF2, KAT2B, POLD3, NMI, IL18R1, NDC80, RNF8, ULK1, RAD21, TACC1, TAGLN2, CFLAR, GPRC5A, PROCR, TBX1, SASH1, SREBF1, RAD52, TRIM21, SND1, NR1H2, SMR3B, TGFB2, PTPN12, MFN2, SHC1, XIST, LRIG1, ST8SIA1, SIPA1, WNT7B, ISG15, WDR5, SETDB1, OPN1LW, WEE1, UCP2, VAV2, RAB31, QSOX1, PTN, UTRN, UGT2B17, SRSF3, YY1, SPG7, TES, CITED2, THRSP, MLRL, AURKB, BCAS2, IL32, KDM2A, SOS1, TRIM13, NR4A3, CHST3, LRPPRC, RALBP1, CCL22, PRDM2, PSMD10, MLLT11, TRIM25, PTPN13, WWP1, PEA15, ID1, MIR148B, FKBP4, FOXF2, FOXC2, FLI1, LY6K, FOLR1, NEUROD1, AZIN2, TP53INP1, ORAI3, FYN, GATA4, GC, MIR449A, TSLP, GHSR, PRKCD, KRT88P, RERG, PIWIL2, GNRH1, NM, MRGPRX4, GSTO2, NUCB2, MED19, BTG1, E2F2, CYP4Z2P, P2RY2, ELANE, PRIMA1, EPHX1, UIMC1, GPR151, MIR381, EYA2, NTRK3, FABP3, NRF1, FANCC, FAP, FAT1, KLF17, FCGR3A, GNRHR, NR0B1, BEX2, KRT8P3, GALNT14, MB, DHRS11, ASAH1, IGFBP4, SLC2A4RG, MIR455, CDCP1, IL7, ARR3, MECP2, RHBDF1, MT1IP, IRAK1, ISG20, PNPLA2, CXADRP1, ITK, ANKRD36B, AICDA, HSD11B2, PBK, AVP, HDGF, SLX4, MIR409, MIR485, MIR146B, H2AZ1, SOX7, ZNF654, MT1X, MT1M, MT1G, ATF4, SHARPIN, MST1R, ALOX5, TRPV6, MIR494, HLA-DRB1, NR4A1, HOXA1, ZNF703, DPP4, MRGPRX3, GATAD2B, MIR19A, CSNK1E, CD80, HUNK, FLVCR1, MIR211, KLK14, LOC111589215, LAMTOR2, VN1R17P, HSD17B13, CTSK, IL22, CTSS, CXADR, GADL1, RGCC, CRYZ, MIR23A, EFEMP2, CETN1, CALM3, MIR107, CALM2, MIRLET7C, COMP, POLG, CALCA, EEF2K, MIR98, MIR99A, PPARD, CLDN3, CDH11, CRMP1, SERPINF1, CGA, DECR1, CAV2, PDGFRA, SDF4, MRGPRX1, PC, BRINP1, PRKAR1A, GPR166P, CBR1, MIR23B, CYP2C8, CCL27, HOXA10, MIR192, EBAG9, CAMKMT, MIR519D, HPN, KPNA2, LAG3, ZFHX3, KIFC1, PPP1R13L, KLF8, KLHL1, FOXA2, PRDM14, CDK3, CDK8, MIR655, SSX2, RGS6, CEBPA, MIR638, MTUS1, MAP1LC3B, HLA-C, MIR7-2, SELP, XK, HLA-DOA, EBP, SRY, MIR193A, NMU, HSD17B2, MIR7-1, MS4A1, USP14, IL25, FZD1, ITGB4, SLC16A4, ITGA2, PIM2, RPP14, PRMT2, KRIT1, CD14, SLC6A4, CD9, MIR215, RHOC, EIF3E, TBPL1, SLCO1A2, MIR7-3, MIR296, ABCG1, GALNT6, APOB, SGK1, HSPA8, HYAL1, CD40LG, IARS1, CD33, SCUBE2, SLPI, HAMP, AQP3, AQP5, JAK1, IL1R1, DMBT1, MIR144, BID, KIF14, CYP27A1, EYA1, CYP2C9, F5, TIAM1, THY1, FANCA, CYLD, THRA, ACSL4, AKR1A1, FBLN2, FBP1, FGFR3, SCGB1A1, TFF2, SDCBP, BCL3, CDK2AP2, MSLN, SCGB1D2, EPHB6, DMP1, HDAC5, MIR135B, BCL2L11, NPNT, E2F3, DDT, FSIP1, ECM1, LYPD5, S1PR1, EDN1, AMOT, EPHA10, DST, EPHA1, MIR374A, EPHA5, PTP4A2, DCD, PWAR4, CST3, CTHRC1, SNORD14B, VDAC1, GCLC, TACR1, MAP3K8, AZGP1, UBASH3B, VHL, GOT2, TSC1, SNORD35B, ECRG4, MIEN1, MIR106A, GSTA1, MDC1, MIR202, CLCN3, HDAC2, SNORD14C, SNORD14D, SNORD14E, CLEC10A, TFAM, FMR1, UGT8, TERC, USP4, PCLAF, VAV3, SAT2, GATA2, B2M, SEMA4D, CRH, VAV1, IL33, KAT5, CRABP2, LIN28B, GFRA1, KLF6, GPC3, CCN4, PTPRO, ZBTB7A, TNFRSF10D, SIN3A, MIR190B, PTPRJ, POT1, PAX6, PTX3, RPS27, PAX5, RRS1, ARHGEF7, SYCE1L, TNFRSF10A, PAK2, PCSK6, PTPRA, MIR708, GNL3, PTGER4, SAI1, PLCG1, PHLPP2, PTBP1, KDM4C, AKR1C3, MCM2, ADAM15, SCGB2A1, CDK19, DELEC1, PDGFC, PGF, ASAP1, RSU1, PAG1, MUC4, PA2G4, RMDN3, KLRC4-KLRK1, ARID4B, TMX2-CTNND1, CDK12, BCAS3, SH3BP4, DENR, NEK3, FBLIM1, NKILA, CORO1C, PLAAT4, ANLN, PRAME, SEC14L2, RASGRF1, CCN3, ALKBH5, GINS2, DSCAM-AS1, NSD3, RORA, SIRT7, SNORD15A, NCSTN, BRD2, PINX1, CEACAM6, ACTN4, NCS1, SF3B1, ANGPTL2, GABARAPL1, SF3B6, SEMA4C, MAGI2-AS3, SMAD1, SPRY4-IT1, LATS2, TACSTD2, MUC5B, NLRP1, PPIA, H3P8, TPX2, DANCR, ZHX2, WASL, PMP22, POLB, KLRK1, TBK1, SMURF1, FOXD3, RHBDD2, SALL4, CHPT1, NUMB, LPO, F2RL3, PRKCZ, LGMN, DYNLL1, EDIL3, MACROD1, ATAD2, PRDM1, XPA, XRCC5, RNF41, TNK2, OXTR, ELK1, TPM1, MIR339, MKNK1, NXT1, ZFP36L1, PDIK1L, RAB27B, P2RX7, LINC01638, EEF1A2, CCDC6, EEF1B2P2, AGR3, CDK5, RAB27A, POMC, NRP2, MIR338, TNKS, RMDN2, PLB1, TOB1, TLR5, NUPR1, CD63, PPP1R1A, TPTEP2-CSNK1E, MIR216A, SOCS2, MIR224, MIR378A, HYAL2, SOX18, FANCF, RABGEF1, NPM1, PGR-AS1, CHD1L, BHLHE22, PRG2, LGR5, FGF13, RBMS3, PIMREG, MIR382, SIGMAR1, SERPINA3, RAP1A, MAPK9, EIF2AK2, MAP2K5, MIR190A, BRI3BP, FLOT1, MIR212, TSPAN13, EPS8, ERN1, MZF1, PPP3CA, BGLAP, ABCC4, H3P17, NTSR1, STUB1, DDR2, F2, MIR326, XBP1P1, TPO, UBE2N, PRPF31, CSK, MTSS1, KDM4A, VTN, SLC40A1, UGT2B15, CSPG4, PIK3R2, PI3, CCR6, WAS, ADAM9, CTSO, MIR134, PRRX1, MIR136, MNS16A, TNFRSF6B, CSH1, RNF144A, UQCRFS1, PTPN22, ROCK2, FZD5, AOC1, PLOD2, LINC00511, COX11, CPB2, PLCD1, PSMD8, ADAM23, OSGIN1, SERTAD1, NOC2L, CA2, RACGAP1, PLA2G1B, KMT2B, CSE1L, FFAR4, PTPN2, RNF168, LOC110806263, CFTR, DLD, CERS6, ABCA1, SARDH, MIR181C, LETMD1, DNASE1, DPEP1, CHAF1A, DCDC2, TRH, TRAF6, ABCC5, BUB1B, H3P9, MPEG1, CDO1, DACT2, MYLIP, ZFAS1, CGB3, TMED10P1, ENPP2, BPIFA4P, CHUK, TWIST2, CD55, EIF2AK3, WNT2, ENPP1, DAP, RCC1, TYK2, VPS51, RMDN1, WDR1, LOC107228383, ANKRD11, SLC13A5, BRD7, TRIM59, LDLR, KDM5A, MAPK12, RTL10, FASTK, MIR483, MAZ, ADIPOR2, BAG3, BHLHE41, CUL1, STIP1, IDUA, MIR484, MIR486-1, RRP1B, TMED10, SPHK2, CUL3, MRPL36, MIR671, KIF2C, IL6R, TLK2, KLK11, FOXK2, HSP90AB1, LYVE1, HSD17B4, HOXA9, SETD7, ITIH5, SLC19A3, SRD5A2, PTGES3, RPS14, HNRNPA1, MIR495, SYBU, MTHFD2, DDIT4, MYDGF, RAB11FIP1, ASRGL1, KITLG, HOXC8, NAA10, HOXD@, HPR, HDAC8, CD46, VASH2, CSMD1, MARCKS, SRPK1, FZD7, CXCL16, ANPEP, PTENP1, CCL8, LMO2, KLK1, KLK2, DDX20, CCL17, CEMIP, MIR605, MIR663A, KRT17, LALBA, MIR608, PADI2, MIR618, SELPLG, LIF, RPSA, MIR630, LBR, SPOP, SLC13A2, KCNMA1, SHH, SIRT2, SLC6A8, AKR1B10, SLC6A2, DAAM1, NUS1P3, PCBP4, IRF6, LTF, SCN5A, ISL1, SLC1A5, KCNJ3, GOPC, RGN, ITGAV, ITGB2, PAK5, LRP6, SIM2, ST3GAL1, MIR542, MIR421, HLA-DQB1, SPINT1, MIR302A, PRDX2, ESRP1, WRAP53, MASTL, ATAD1, SLC34A2, MRPL28, AFAP1-AS1, TDO2, CBX2, EPSTI1, EGLN2, NELL1, SPZ1, KISS1R, ST6GALNAC2, PRMT6, SPAG5, CYTOR, ABCE1, CDK5R1, DOT1L, ANP32B, RPS6KA5, CBX5, SEMA3C, MIR448, BAK1, MTA2, BCAS1, NCAM1, ITGB3BP, CGB8, PES1, KSR1, TAT, GJB2, CGB5, GAST, RNASEL, HOTTIP, NEFL, MIR452, GPX3, MYCBPAP, QRSL1, C20orf181, NUS1, ATP6, LARP6, ALOX12, ST6GALNAC5, HIVEP1, MSR1, EGLN1, CHDH, NME2, CD200, SAGE1, MIR493, MMP17, RBBP7, NFKB2, NGFR, CREB3, SCRIB, ANTXR1, MTTP, CYGB, MUC2, KLLN, MIR425, NLRP3, MIR1246, GSTM3, TIMM17A, MAP3K14, MIR34C, PRB2, CCAT2, MIR544A, GATD3B, TRIP10, MIR660, MIR30E, MIR379, MBTPS1, MIR661, HNRNPA1P10, ANGPTL1, PANDAR, MIR361, MIR564, NOL3, CDR1-AS, SART1, TNFRSF10C, CISD2, UBE2L6, MIR506, MIR502, FHL5, TRAP, RAB11A, MIR324, MIR876, MIR216B, DDX21, MIR940, MIR526B, RN7SL263P, MIR944, MIR492, MCM3AP, MAP4K4, MIR133B, AIM2, MIR760, POTEKP, CBFA2T2, MIAT, XRCC6P5, POTEF, SNHG14, MIR323B, FGF18, MIR372, MIR370, SNHG6, POTEM, LINC02605, MIR330, MIR1271, WASF1, MIR1258, BLID, TEC, APLN, TRIM24, MIR300, TNFSF13, GLS2, PTGES, TCIM, TMPRSS4, SUSD2, CHFR, ENAH, TENM3, NSUN5, HIF1AN, STAB1, AMBRA1, FERMT1, HJURP, SLC39A9, BRF2, MGA, IMP3, NAT10, TRIM62, SATB2, CEP55, CDCA8, PPP1R13B, KIF26B, RNF31, AKIP1, PMEPA1, MIR25, SMG1, WDHD1, LZTS1, PLEKHB1, WIF1, AKAP13, ZBTB4, ZNF398, MIB1, PARK7, ACOT7, OIP5, CASC3, AHRR, RRAS2, ITGA11, RAB22A, CBX8, ARFGEF3, MLXIP, TP73-AS1, TWSG1, CIAPIN1, CEACAM19, TUG1, KCNH4, CT55, CYP2W1, KCNK9, GLCE, ASCC1, SOST, IL20, CHST11, NOX4, KLK12, SH3KBP1, NOP53, FBXO22, PSMC3IP, HSPB8, SENP1, CARD10, SNORD44, RNF11, DNAJC2, UBE2T, GIT1, TMEM97, POLL, CACYBP, UBR5, NELFB, GTSE1, PTOV1, CASZ1, NSUN2, TRIM44, LEPROT, PRMT7, AMACR, WNT4, ZMYND8, ERRFI1, FXYD5, TMEFF2, CXXC5, SHC3, RSF1, RTRAF, LSR, SUN2, UFM1, ACKR4, QPCT, VTA1, SULT4A1, NTN4, TRPV4, SENP2, TMEM132D, PDCD6IP, PARP3, MARCHF8, SLC25A43, PRSS55, PTPRU, ACTR2, BCAP31, CNTN4, IL23R, G3BP1, C1orf52, LYPD4, LINC00052, BCDIN3D, PSME3, CACUL1, FOXR2, SPRY1, TAAR1, FSTL3, KCNH8, ACVR1C, IL27, CASC2, NR2E3, DCAF1, PRDX6, MIR199B, MIR196A2, MIR188, MIR154, MIR130A, ST18, MIRLET7G, RAB11FIP3, C4orf3, MIR100HG, TINCR, TMEM189-UBE2V1, TMEM189, MAML1, NDUFS7, MACC1, ACTBL2, SCARA5, HOXA-AS2, HS3ST2, NR1H4, MSI2, C10orf90, ERLIN2, TUBA1B, PEAK1, NOL9, BRD8, FAT4, SPIN1, WDR77, TRPM8, AHNAK, MARCKSL1, TUT1, TNS3, TMPRSS3, RIPK3, COP1, ADRM1, NOD2, LRRC4, TNN, MIIP, KAT7, RINT1, AFAP1, PBOV1, LPCAT1, LINC00472, DHDDS, HOPX, KLHDC7B, CRB3, NAF1, CREB3L1, CIB2, PYGO2, WNT3A, SLC22A16, HTATIP2, SNHG7, PDPN, ZC3H12A, IGF2BP1, RPAIN, RHBDD1, IGF2BP3, IGF2BP2, VMP1, CLDN16, HM13, PDCD1LG2, PDGFD, SLURP1, H3P40, HNRNPM, MYO10, E2F5, NOS1, NONO, NOP2, TNFAIP1, EFNA5, EFNB2, DRG1, AMPH, TPI1, EIF4A1, CRISP2, TTC4, ELAVL1, MYLK, CCT3, MXI1, MVD, TRPC1, ELN, TRIM37, TRPC5, MARK2, EMP1, DVL1, NUMA1, SLC25A5, OPRM1, TCF3, DDB2, SLC25A3, PGK1, TRA, CDK14, PF4, GSDME, TERF1, DKC1, NR2F2, TFDP1, DOCK1, TGFB3, PBX1, PAX2, DRD2, P4HB, TIE1, ANXA7, TIMP4, ANXA4, TK1, MTNR1B, COX1, PDHX, LGALS8, FES, XPO1, MXD1, TM4SF1, SH2D1A, LYN, LPP, FKBP1A, LPL, LIMS1, SLC30A1, LGALS3BP, ALPP, AFF3, LAD1, PAX8, KRT7, TFEB, SEM1, KIR3DL1, AP2A1, FRK, PSCA, FRZB, FECH, FCGR3B, FCGR2A, MAS1, TTN, MPST, MOS, EPHA4, TYRP1, EPHB1, AFDN, ALDH2, UBE2V1, UBE3A, MEOX1, SUMO1, UGT1A, AIF1, ETV5, EXTL3, USF1, MDK, F8, MCM7, MC4R, TRPV1, VRK1, DCC, PLAUR, TAP1, BHMT, SEMA3F, RORC, SELENOP, RNH1, RNF5, BMPR1B, BMPR1A, RNF2, FOXN3, SGCG, RGS4, RFC2, DAB2, BCYRN1, SIAH1, ST6GAL1, RELB, REG1A, SKI, RBBP6, CCR4, SLC2A3, ACKR2, SLC3A2, RPA1, RPA2, SDHB, CDK11B, SALL1, CASP2, CASP10, CBFA2T3, S100A11, SORT1, SLC25A20, CCL1, CD1D, CCL4, RPS6KA2, RPN2, PTTG1IP, C5AR1, CD28, C5, CCL19, TNFRSF8, XCL1, CD38, RPL17, C1QA, CD70, BCAT1, RAN, SLC12A3, HTRA1, RELN, PROX1, SQLE, ARSA, SRF, CTBP2, CTH, SRGN, PPP2R2B, ST13, ARHGAP1, PPP2R1B, PPP1R8, CYP2A6, TRIM23, PODXL, ABCC8, SUV39H1, PMAIP1, CYP26A1, AQP1, SYP, TAL1, KLK7, PSAP, COL5A1, CSH2, RAD9A, RAB5A, ATP7A, RAB1A, PYCR1, ALDH7A1, CP, SMARCE1, PTPRF, SMO, CPD, SNCB, PTPN9, CPT2, CR1, ATIC, PTGIS, PTGFR, PSMD4, SOD3, SOX5, CRKL, CRY1, KCNH2, CAPN1, SEMA7A, HMGN2, HLA-B, ADA, ACTG2, IGSF1, ACR, FFAR1, ADAM8, PARP4, GTF2H4, HNRNPU, GEM, IFI16, HMGB2, HMGB3, OGT, PIAS1, IRF5, STK24, GSTZ1, GATD3A, GBP2, CDC7, GCG, GATA1, GNB3, GDF1, ACTG1, IDH2, HDC, HSD3B1, IRF3, HTT, SLC7A5, IFIT3, USP11, HOXC10, GLB1, ING2, IL5, HOXA@, HNRNPC, BRF1, PPFIA1, GUSB, MAD1L1, GYPA, KAT2A, ITPR3, GLI3, FTH1, IL9, ITGB6, GSTA2, AXIN1, ACP3, AANAT, HOXC11, IL13RA2, HSPD1, PDE5A, RBM10, GTF3A, IL3, GPD1, MAPK11, HCAR1, MAPK6, CST1, PRAC2, CSRP2, PALD1, CSTA, WLS, PDE7B, LINC01193, TUBA4B, HCN1, SKA2, FOXD2-AS1, DKK2, STEAP1, DNAJC3, KDM2B, TAS2R38, CCL4L1, GLYR1, PSMD3, FBXO10, FBXO5, ERAL1, HOXA7, PSMC3, CRIP2, HOXA11, LHX6, PREX2, HOXB@, AATF, SIGLEC7, CSHL1, CKAP2, HAVCR1, CSN1S1, SPSB1, HBP1, LINC01116, HOXB3, CSNK2A1, HOXB5, NPTN, MLH3, PIF1, NCR3LG1, SOX2-OT, FSHR, LSM1, PADI1, DOK7, UBQLN1, A1CF, RGMB, HSPA9, PLCB2, DAPK3, PLCG2, DBT, DAG1, SCAI, RMC1, PLD1, E2F8, LIN9, PLEK, PLAGL1, ZNF326, HSPA2, FBXL19-AS1, BCL9L, CENPU, AKR1C1, PGC, FGF14-AS2, GLP1R, PGP, GLUD1, PKD2, MIR22HG, CXCL17, KLK9, DCK, ADGRE2, GLUL, RSPO1, ICOS, FXYD3, PDLIM3, GOLGA2, PPP1R2, HPRT1, SLC27A6, DCPS, CYC1, HPD, PPP2R2A, CYBA, PPM1B, ABCB5, HTRA2, CPP, PRCP, PREP, BMP10, CRCP, LINC01139, PPIB, POTED, GNAS, PLP2, GNA11, PARVB, PLXNA1, SEPTIN4, GNA12, KDM8, POLE, PPBP, FHOD1, POR, DNAJC15, DROSHA, PIP4K2C, CLEC4D, LINC00339, PHF5A, SMIM10L2A, KLK13, MIR199A1, KIF18A, GTF2H2, USP22, MIR196A1, SIN3B, CDC6, MIR198, MIR199A2, CDKN2D, RPL10, HCLS1, ANKRD12, CD81, HBB, CD69, HAS1, RORB, CDR1, CEP70, SIRT5, CHI3L2, ING5, CHD4, RIEG2, MIR15B, PDIA3, SIRT4, GSPT1, MIR187, MINDY4, MIR181A2, RAB1B, CENPA, MIR184, LOXL4, MIR186, ASCC2, AIF1L, HABP2, CBS, PHF8, CCNG2, CCNG1, H1-0, CCNF, ITCH, CCNC, S100A10, MIR208A, ROPN1L, EMILIN2, MIR30D, SARS1, CAST, PALLD, MIR32, KDM6B, CCT6A, SEPTIN6, MPRIP, FBXO28, FAM107B, SULF1, KAT8, SESN2, RHNO1, ACSBG1, CD6, KANK1, FERMT3, RCOR1, MIR219A1, CRISPLD2, TLCD3B, RPS15A, AKT1S1, CHIT1, RFX1, PGLS, POC1A, NECTIN1, MAK16, BACE2, GPR39, MTO1, RAB2A, RAD54B, RAD23B, WNT5B, RAD17, PANX1, ZBP1, KIF4A, BRD1, FLRT2, MR1, RCHY1, SOSTDC1, PTPN23, PTH1R, PTPN3, ATRNL1, TNKS2, HNRNPA2B1, CSAG3, TENM4, TIPARP, USP17L9P, KANK2, PNKD, AKR1C2, HNMT, AFAP1L2, CPA1, MIRLET7E, COL4A2, RFC3, SUZ12, PSD4, DNPEP, RBFOX2, RBL2, RBM4, SRRM2, RBP2, MNX1, PLPP5, SNX27, FAM83D, ANP32E, CBX7, REV3L, ELOF1, CHRNA4, MIR130B, GRB14, MIR129-2, DDAH1, HLA-DQA1, MIRLET7I, SMPX, RAP1GDS1, LCOR, PLA2G15, CNR1, SNHG1, CMKLR1, GRK5, RARRES2, RASA2, DDX58, BRSK1, TRIM11, PFN2, CYB5R3, TMED5, ZNF395, INS, TSHZ2, FBN1, FBRS, CMPK2, RBM45, ACSL1, NSD1, F10, PCDHGB7, ALG1, MCM6, F9, ARHGAP9, F7, CPEB2, ACSS2, METTL3, LTB4R2, CCL28, PNO1, MAGEA2, GARS1, LXN, TINAGL1, SMARCAD1, TNFAIP8L1, SEMA4A, OSCP1, IQGAP3, MAN1A1, ROBO3, WDR20, LINC00628, TSACC, BCCIP, DIABLO, UNC45A, GAS6, PFKM, ECHDC1, MGAT5, IL16, ERG, DOCK11, S100A16, MIP, EPS15, DYNLL2, MAP3K9, DEPDC1, MAP3K10, ASAP3, MAP3K11, IL15RA, UBE2Q1, CHRNA9, ILRUN, ATF7IP, MGAT3, TNFRSF9, ILF3, ANKRD44, MAP3K3, GBP1, MAP3K5, ST6GALNAC1, ETV1, ETFA, CMTR2, MEST, TDP1, PRR11, ETF1, PDIA2, OGDHL, AGK, CCAR1, TMEM88, SMYD2, MEPE, FGF5, FOSL2, ITGB5, KIF11, FMOD, MYOCD, MIR155HG, PHRF1, CARD16, USP28, KRT6A, ITGA4, STIM2, KRT13, CFAP97, L1CAM, SRR, SMYD4, LACTB, KNG1, UCN3, FUT1, EGLN3, ACE2, IL21, KCNN3, SLC25A19, FRA16D, PRUNE1, KIF2A, KIR3DL2, FRA7G, NGB, PTBP2, CYP4F11, MARK4, FOXP2, KIR2DS1, PIEZO2, ITGA2B, RPTOR, LY6E, FH, FGR, LTB, LRRK2, LTBP3, H4-16, BCAM, PDXP, PDX1, TIMM50, INSRR, ADAMTS9, TPPP2, IFI6, TC2N, PAQR4, LRP5, ACKR1, ARHGAP31, TENM2, LAMC2, ARL11, KIDINS220, LCN1, FOXJ1, FUT8, LGALS4, IRF7, LOXL1, LGALS9, LIG3, PBXIP1, UBE2Q2, VANGL2, EIF3H, VWA5A, ENGASE, E2F7, MNT, EPB41L4A-DT, PTRH2, OLR1, GFAP, GFPT1, CT83, BRCC3, SLC36A1, TSPYL5, NOX5, OXA1L, PAGR1, PAM, NCKIPSD, ID4, NOP16, ISYNA1, NKD1, DERL1, HBEGF, OAS3, SYNPO2, CITED4, CHAC1, TERF2IP, NPPA, DDX53, LINC00160, DUSP6, ADAM22, CMPK1, SPA17, DUSP2, MAP3K20, NTS, MAPKAP1, TMEM8B, CADM4, MRPL58, PRRX2, ADGRF5, ARHGAP10, NUSAP1, CPA4, PDC, PCSK9, HRH4, SLC26A4, RHBDF2, MLXIPL, TFDP3, GAL, NDUFA13, MEMO1, MAGEA2B, ISOC1, DHX9, DDX1, RICTOR, GOLM1, PCDH7, HOXA11-AS, KDM1B, SERPINA5, ATRAID, POMP, LINC00518, CHST15, PCP4, GJA3, HTR2A, FZR1, TNFAIP8L2, GJA8, TLR8, SPDYA, PDB1, DNAH8, FGD3, NNMT, SMPD3, NMT1, TBL1Y, APPL2, MUC6, LINC00473, IL4R, CASD1, MCU, MSTO1, MYCL, MYH2, MRPL41, MYH10, MYH11, FANCL, PRPF38B, ELF1, IL2RB, IL7R, EMP2, ASXL2, MSX1, MPG, HES6, ZNF331, MCM10, MPZ, CITED1, MSRA, SYNJ2BP, MTHFD1, CERK, DRAM1, MSX2, MTAP, ATP8, P2RY12, NUDT1, SERPINB1, GPAT2, GPBAR1, EFNA2, NEDD8, ZBTB10, PEBP4, NEO1, ERCC6L, RHPN1-AS1, PPP1R12C, EEF1A1, FNBP1L, SLC5A8, NFYA, SLC25A21, DNAAF4, ECHS1, NNAT, UGT1A8, PGPEP1, CELSR2, LAMTOR1, EGR3, RBPJ, TESC, CAPN2, NAIP, NAP1L1, IGKC, ODAM, SARS2, SEPTIN2, PAQR3, MLIP, KLB, NDN, FAM53A, NDUFB3, SLC2A12, VAMP8, PTGDS, SLC14A2, MIR1301, HIF1A-AS2, PDLIM1, ATP6AP2, PDZK1IP1, AFM, SSTR5, SSTR1, SSRP1, ARHGDIB, SMIM10L2B, WNT11, MIR1290, TNFSF13B, AIFM1, SSAV1, TNFAIP3, CTR9, CRNDE, USP39, NRG3, ARNT, USP8, CERNA2, ARRB1, ARRB2, ANG, H3P12, LPAR6, NME1-NME2, ARID3B, OSMR, ARF3, MYOM2, HEXIM1, TMPO, COX7A2L, PCSK7, RBM5, RBCK1, MIR4732, TKTL1, EPX, TRIM16, CELF2, ZFX, SULT1A2, AURKC, ZNF197, H4C9, STK3, STAT2, STAR, LINC00598, ADRA2A, ACOX2, LINC00461, PLK4, WNT3, SIX2, MIR454, SOX11, BIN1, SPAG9, MIR500A, SOX3, WIPF1, MIR668, HCP5, ASPH, PKD2L1, ARPC2, LSINCT5, SNTA1, RAI2, MIR520C, BLACAT1, TSPAN1, MIR1204, PRNCR1, ATHS, HAP1, MALT1, MIR498, ATOX1, ARPC1B, CERT1, TPD52L2, FRY, AGTR2, SPAM1, RAI1, H4C4, CLIP2, CHL1, GIPC1, SPRR2A, H4C6, H4C12, TNP1, H4C11, CAPG, H4C3, APOBEC3A_B, NBAT1, OPTN, LINC01133, CCR9, TRAP1, ANCR, H4C2, STARD10, H4C5, H4C13, MIR505, H4C14, DVL1P1, SULT2A1, BLCAP, TLE1, PEMT, KAT6A, PIAS3, TFAP2B, RECQL4, EEF1E1, RECQL5, VPS4B, GOSR1, LIPG, BRD3, ZFYVE9, NCOA4, RACK1, IFITM3, RAPGEF3, LINC00673, SSX2B, SLC25A16, RBM14, MAP3K12, LONP1, FAM3C, TMBIM6, ADAMTS9-AS2, TEAD4, SELENOF, TFCP2, MIR539, LUCAT1, MAEA, ATG5, MIR652, DLEU1, ST8SIA4, KLF10, SLC39A7, AP2B1, MAFK, THBS2, TLR6, APBB1, KLF2, MIR3646, MICU1, TGFBR3, NKX2-1, NTN1, TUSC3, AIRE, TUBB4B, TKT, SYCP2, DNALI1, CNKSR1, NOD1, NBR2, SLC9A3R2, MAD2L2, MIA, TBXA2R, MIR4728, P3H3, DLG5, DLGAP1, CDR3, GLRX3, KIR2DS2, TARBP2, PRDX4, ARL6IP5, TAP2, APOD, PIBF1, SLC19A2, MIR411, DCLK1, OLFM4, AKAP3, RBM39, CCT4, CCT2, SLC33A1, KIF20B, AQP4, H3P41, NOG, HYOU1, ZNF32, ZNF35, MIR597, EI24, MIR592, TADA3, NXF1, GRK2, TRIP13, TCF21, LINC00968, RBM14-RBM4, RPL17-C18orf32, ADRB3, MIR574, ADRB1, TCL1A, CCL4L2, ZNF143, MIR569, TCF7, MIR568, FBLN5, ELOC, APOC3, EIF1, GDF3, SNCA, H4C8, ACYP2, ACP1, MIR874, API5, LOC100288966, FGF19, UPP1, HPS5, MIR346, SELENOW, SIRT1-AS, ACP5, BNIP3L, KHSRP, SLC23A2, MIR422A, MIR331, TUBG1, SEL1L, ALOX15, TUFT1, SOX2-SRR2, LINP1, MIR328, KLF9, TUSC2, CBX3, UGT2B4, KLK8, TUB, UTS2, HCAR3, MIR384, TOX, LINC01857, BVES, PARG, RASSF2, CORO1A, PMEL, ACTN1, PTBP3, NUDT5, IKBKG, IFITM1, SHMT2, BDKRB1, CFB, HNRNPDL, SH3GL1, THOC1, IRAIN, GACAT3, CBX4, MIR543, MIR937, POLI, GABARAP, UGDH, MGLL, CCL7, UBC, TNFSF9, LNCRNA-ATB, SRL, TNFSF12, TNFSF12-TNFSF13, MAPRE1, PLA2R1, H3P7, GPAA1, SNW1, UBE2B, KIN, H3P24, AGAP2-AS1, H3P42, PTGR1, S1PR4, UBE2E2, H3P47, MIR9-1, ALDH3A1, CAMK2B, NINL, CANX, CA5A, MAPT-AS1, MIR302B, MORC2, SPATA2, MIR151A, TYR, SDC4, SDC2, CX3CL1, EGOT, TYRO3, C3, ACLY, MANCR, ZNF652, CCL25, IQSEC1, PLPP1, CCL16, USO1, DLK1, ST8SIA6-AS1, FADD, RIPK2, MIR2052HG, HSD17B6, CA1, GINS1, SIX3, THPO, SART3, MIR888, MIR675, AK6, MIR511, SLC7A1, H4C15, ATP2B4, VGF, ATP2B3, CACNA1H, CNMD, SMARCA5, CBX1, CACNA1I, MIR765, AMD1, PRDX3, TOPBP1, BCL2L10, MORF4L1, DNAJB4, MIR362, VTRNA2-1, MIR665, MIR1247, MIR410, AMHR2, IATPR, BTRC, RANBP9, AZU1, HSP90B2P, ATP6V1C1, FAF1, KIAA0100, PARP2, SMARCA1, MIR433, BCL10, SLCO2A1, SLC25A1, MIR491, MIR18B, TRPM2-AS, BAAT, TRPC3, ZGLP1, ATP2A2, MIR1307, MIR301B, SELENBP1, EHMT2, LIMD1, ATP1B2, RAE1, USP1, MIR4435-2HG, MIR873, RCAN3, CAVIN2, CSAG2, ABI1, SLC2A4, GHET1, TPM2, SMS, SMPD2, SETD1A, TRPM2, ACTRT1, MIR1229, TAF8, MIR1266, MIR3666, LRG1, MIR3908, MIR3617, F11, TGM7, CNTROB, TOP1MT, MIR1207, PNCK, FANCB, FAH, ZFHX4-AS1, FOXL1, FANCE, MIR3942, MIR1256, GRIN3B, MLANA, MIR1296, ZNF501, ATG4A, AD11, FOXS1, ALAD, TNFRSF13C, MIR3609, MIR3614, FLG, FUNDC1, UPRT, DBH-AS1, ERV3-1, MIR3922, FBN2, PIGU, MIR3923, MIR1297, UHRF2, FBXO32, MIR1228, FLII, JMJD7, OMA1, FOXE1, BHLHE23, WDR92, AHCY, AK4, PIFO, VASN, MAGEE2, AFA1, MIR374C, H3P20, UHMK1, FOXD1, CPT1C, MIR548J, MIR1471, FGA, AEBP1, EVI2B, TDRD9, LOC100422227, SPPL3, MIR1267, MIR3196, MIR1915, MIR1469, MIR1236, MIR3188, TDRD10, NCOA7, MIR1179, HMGB4, MIR761, TSPY10, MIR2276, FGF9, EMB, USP43, ZEB2-AS1, TMEM132E, MIR663B, F2RL2, EZH1, CYB5D2, MIR762, GNG12-AS1, MIR1284, FGD1, EXT1, TBC1D16, TMEM219, EWSR1, SETD9, MTPN, LOXL1-AS1, MIR1193, GOLT1A, LINC00899, FKBP1AP1, AHSG, MIR1972-2, MIR3141, MIR4262, MIR3184, FKBP1AP2, MIR4282, FCER1A, MIR1249, PXDNL, FKBP1AP3, FKBP1AP4, CD200R1, FOXG1, THEM4, ANTXR2, MIR3065, ALKBH2, MIR1245A, ETV3, FGD4, VPS37A, MIR4293, MIR1287, FGL1, MEF2B, MIR4316, MIR664A, SFR1, FCGR1B, MIR4301, MMP21, FHL1, FHL3, EXOSC6, MIR3200, FOXN4, MIR33A, RHPN1, KLHL22, C3orf67-AS1, MIR503HG, ZNF496, ABHD14B, PLCD4, ADAMTS9-AS1, ODC1-DT, GPAT3, LINC01585, LOC101930370, KMT5C, GOT1, SPSB2, GPLD1, GPM6B, GPT2, LOXL3, MIR6826, CCDC54, PPP1R9B, CCR10, SLC9A7, MIR6861, MIR6882, TRIM63, MIR8073, MIR7847, USP32, MIR6875, ZNF503, RIOX2, COMETT, GOLGA1, FOXD3-AS1, RGS8, GHRHR, GIP, LINC01355, NPY4R2, GLA, MYLK2, ALG2, KRT90P, ADH5, PLIN2, SNHG12, FAM83A, EZR-AS1, ADCYAP1R1, ATG4D, ADARB1, GM2A, MAEL, ATG4C, PPP1R15B, GNAQ, FBH1, LINC01483, LINC01787, GNB1, ACVRL1, PTPN5, MIR8084, XCR1, ROCR, USP38, GSR, SNORD138, GSTM4, TMEM126A, TRAF7, GSTM5, LOC110283621, LOC110599580, ACAT2, ACADSB, BTG3-AS1, ABR, KIAA1109, RNASEH2C, SIRLNT, ABL2, GTF2I, ABCB7, AARS1, AAMP, USP44, H3P28, RAB6C, GUCY1B1, MAGT1, AADAC, H3P33, GYPB, CCDC8, HSDL2, TALAM1, GRM4, GRK4, GPR17, LOC102723971, ACVR2B, LNCTAM34A, GPR19, ACVR2A, ST6GAL2, SLC12A8, MAP1LC3A, GPR35, LINC01287, FOXC2-AS1, MCM8, PERCC1, GRIN1, LOC105371849, MAP3K21, SYVN1, MKNK2, GPT, ACTC1, GPX7, LINC01614, LOC105377871, ACTA1, LOC108004545, HVCN1, SARNP, NKD2, B4GALT1, FLT3, CENPI, MIR497HG, BIRC8, GAS1RR, TUNAR, CCAT1, FTH1P3, FTL, ABCC12, FUCA1, ORMDL3, LINC00707, SFXN1, FUT2, CT47A12, SNHG16, ARHGAP18, C8orf17, FUT3, LINC00458, FUT6, PGAP3, LINC01016, LY75-CD302, ZNF276, MAFG-DT, BCL2L2-PABPN1, RAB4B-EGLN2, ATXN3L, SPECC1, CAVIN3, MRPS30-DT, DCLK3, IL17F, CCDC85A, RAD51-AS1, UBE2Q2L, ITGB2-AS1, PSMG3-AS1, FLT3LG, LINC02582, FMO5, MAL2, AP2A2, LOC100505909, LINC00963, HOTAIRM1, GAS6-DT, CBR3-AS1, LINC00310, SLC35A4, FPR1, PIK3IP1, FRA6E, LIFR-AS1, CYP2U1, CMTM1, GRK3, TMEM54, FRA7H, KIF12, CDCA5, MTFR2, FYB1, ADRA2C, BIVM-ERCC5, GABRA3, GCSH, ADRA1D, NNT-AS1, GCY, ADORA2B, DUOXA1, MIR5188, BMF, HEIH, MSTN, UNC5A, CERS6-AS1, KCNH7, FSBP, WDR34, CYP3A7-CYP3A51P, AQP10, SEC16B, ERVK-11, ABCC10, NAV1, MYB-AS1, STARD13-AS, HOXC-AS3, HOXC13-AS, ADORA1, IFNG-AS1, SCIN, GFM1, ERVK-22, ERVK-15, ERVK-12, MIR4480, GAD2, TMEM132C, GALNS, ERVK-2, LRRC32, MIR4469, ZNF300, CHRDL1, MIR4428, GAS1, MIR4458, GAS8, SLFN11, DERL3, CARMIL3, GATA6, BOD1, MIR4530, L3MBTL4, MIR4513, MIR4485, GBX2, MIR4766, MIR4665, MIR4484, JPT2, MIR4443, GCNT2, GPR119, FEZF1-AS1, MUC17, NPS, CSNK2A2, ASCL2, HCC, MBOAT4, LINC00589, KLKP1, HAS2-AS1, HDDC3, ENHO, MIR510, SLC27A1, MIR509-1, CA13, MIR507, ASCL1, ARSF, RBMY1D, CSTF1, RBMY2DP, SLC35B2, ARL2, AMIGO2, ARPIN, BCRP1, ARSD, ASPG, NUP43, ZACN, RGSL1, MIR487A, DND1, CSNK2B, ASIP, HMGA1P7, MIR567, LRRC26, PCARE, TSPYL6, MUSTN1, CPM, MIR496, ATP1B1, C1QL3, MIR99AHG, MIR432, BRCA1P1, ATP2B1, CPB1, FENDRR, COX6C, LRRC75B, HRNR, SNHG5, SERPINA13P, ASNS, ASPA, CRIP1, C10orf143, C12orf75, CREM, MIR520G, CRABP1, RTL1, TNFAIP8L3, ASS1, CRAT, IMMP2L, YPEL2, IGFBPL1, CTNNA1, ARHGDIG, AQP7, DAB1, MIR33B, SNORA7B, CELIAC2, CHSY3, CYP11B1, CYP4B1, TRARG1, CYP2J2, PAX8-AS1, CYP2D7, AQP9, S100A7A, ASAH2B, EIF4E3, DGKA, ZFP57, MIR553, STPG4, MIR562, EPHA6, SUMF1, CTBP1-AS, MIR559, LINC00491, MIR449B, MROCKI, MIR552, DAZ1, SNHG15, DLX6-AS1, MIR551A, CT47A7, ARF4, IFNE, CX3CR1, VSIG1, SIAH1P1, NANOS1, NEK5, CYB5A, STAC2, CTSZ, HSR, RSPO4, TMPRSS7, CTSG, CTRL, RASSF10, CTNND2, ARHGAP5, RSPO2, TSPAN33, LINC00520, ARSI, CYLC2, RHOB, LINC00273, C3orf35, ARMH1, CLEC4G, CYP2A7, LINC01089, CYP3A7, HCAR2, SCFV, CYP2C18, COX5B, LINC00668, COX4I1, ADGRE5, CDH4, CDC25B, MIR197, BTF3, SEPTIN7, CDC5L, BYSL, CDH15, ENTPD5, ENTPD3, BPY2C, BPY2B, C4BPA, CD86, BTD, CDH17, CHD1, CETP, MIR16-1, CEACAM8, MIR196B, BMP5, BMP8B, MIR367, CENPE, BTC, CEL, BNIP1, CLGN, BOK, BPHL, ZFP36L2, CD27, GAS8-AS1, CD22, RUNX1T1, MIR95, MIR28, CALML3, CBR3, SERPINH1, CBL, MIR92A2, CD8A, CASP6, CASP5, CASP1, CALU, CAMLG, CAPNS1, CCKBR, CALD1, CAD, CACNA1E, MIR24-1, CCT, CD1A, CA11, PIM3, CD3D, SMIM22, CD3E, MAP1LC3C, MIR218-2, MIR217, CD5L, NRARP, TARP, CHD3, HAGLR, BCAT2, MIR363, MIRLET7D, PGM5-AS1, MIRLET7F1, ANKRD30BL, BAGE, BCL2L1, MIRLET7A2, COL4A1, MIR101-1, CNTN1, CNP, KIR2DL5B, PLK3, COL4A5, HNP1, AKR1C4, CUEDC1, CORT, MIR488, CASC15, KPNA7, IFITM10, COL15A1, LINC01561, MIRLET7A1, MIR376B, ATRX, KIF1A, COL12A1, COL10A1, ANKHD1-EIF4EBP3, CNGB1, LTB4R, MIR124-2, CHM, CIRBP, BCR, CKB, BDH1, MIR377, CHRNA5, EID3, ATXN7L3B, SUMO1P3, BNC1, MIR147A, CHGB, CHGA, POTEE, CKS2, CLCA1, CLK2, CLN3, MIR133A1, TNFRSF17, CLTA, BCL9, CLTC, BCL7A, MIR128-2, BCL2L2, CCR3, ANIB3, DUXAP9, MIR125B1, CT45A1, MIF-AS1, RUSC1-AS1, ASB6, TMTC3, MOSPD2, RHOXF1, USP51, USP17L30, SLC35G1, EDN3, S1PR3, MEMO1P1, CLEC14A, EDA, SPRED1, USP17L29, SLFN5, FBXO39, C2orf92, SHOC1, USP17L28, IQUB, TBC1D3, DAB2IP, IRX2, PRSS3P2, AMOTL1, OIP5-AS1, TEX46, EFNA3, UNC5B-AS1, SPANXB1, ANKRD46, HULC, LRATD2, LINGO2, PLPPR5, CALML6, LOC284454, CT47A1, MIER3, DUSP8, DUSP7, BHLHA15, HTR3C, TSPY3, PSORS1C2, LINC00460, ADAMTS17, ADAMTS18, DUSP5, ASXL1, CT47A2, DNAJB1P1, SGMS2, RASSF6, IFNLR1, HFM1, USP17L27, ECE1, USP17L26, ANK3, USP17L25, USP17L24, CCDC116, MAGED4, E2F6, WBP2NL, DEFA1B, DYRK1A, CARMN, DUSP9, MEGF9, EIF2S1, BTLA, EPB41, MIR939, KRT80, MIR933, EPB41L2, A2ML1, FBXL14, SLC38A6, OR2AG1, ALPI, ENSA, ALPL, MIR942, ENO2, ALX3, MIR365A, TTC36, EIF2B1, NRSN1, FOXO6, RAB6C-AS1, GATA5, ALDH1B1, EPRS1, TFAP2A-AS1, FGF13-AS1, ALOX5AP, SRXN1, SIRPA, MIB2, EPHB3, ABCD2, MUC15, HAPLN3, MIR934, NRG4, EIF4G2, AMELX, TMEM170B, PYHIN1, SHLD1, SIK1, AIFM3, AMPD1, EN2, LINC00309, EIF4B, PSS, MIR802, MIR766, EIF2S3, ELF4, MIR298, AMD1P2, HJV, MIR450B, ELK4, CREB3L4, EMD, SPC24, TMC4, APCDD1, AMBP, SLC25A52, CCBE1, TMEM199, EMP3, IL34, CT47A3, AGO3, AGO4, DLAT, PROSER2, OR2T6, DLG3, ANKK1, CT47A11, MIR612, DHPS, DLX2, DHODH, GPX6, BIRC3, FAM133B, DFFB, APOA4, RNF169, MIR613, FOXK1, RNASEH1, SRRM3, TRDMT1, FAM83B, MAPK15, MIR616, DNM2, NEIL2, BRWD3, DNA2, DNM1, MMS22L, DLX5, MSRB3, YTHDF3, ASPM, MIR603, DFFA, DCX, CASC22, MIR588, MIR584, MIR578, NEK8, MIR577, DCTD, MAGI3, MIR573, ZNF763, WDR62, ZFP82, MIR7-3HG, SLC25A41, DDC, LINC00641, MIR589, MUC19, RASSF3, APOC1P1, MIR591, OR10A4, MIR593, MIR599, MIR600, DEFA1, HCA1, NPAS4, MIR601, HS6ST3, STEAP2, THSD7A, MIR621, PLPP4, FLCN, CRTC2, DSPP, TET3, PIKFYVE, TMEM17, ARL13B, SNORD118, LINC00466, RDM1, DSG3, HTRA4, DSC2, SLC25A6, DSC1, SLC26A2, SLC25A4, DOCK3, CT47A6, IMMP1L, RILPL2, CT47A4, CT47A5, METTL7B, RMST, PAOX, CT47A10, CT47A8, CT47A9, ADCY4, CT62, MLKL, GGN, DRD5, TMEM26, DRD4, DPYSL2, ALKBH3, MIR92B, SKA3, MIR645, ZUP1, OPN5, APAF1, TBATA, DPYS, DPT, MIR628, MIR627, DOK1, MIR622, MIR650, DPYSL3, SLC26A3, FAM171A1, UCMA, JMJD1C, DRD1, HEPACAM, OTUD1, MIR653, DOK6, LRTOMT, DDIAS, DRD3, SLC37A2, ANXA13, HYLS1, SLC4A11, LMNB1, TATDN1, CD160, PLAAT3, HHLA2, ST3GAL3, SHOX2, STRAP, ADAMTS6, FBXW4, MAP4K1, SH3GL2, ITSN1, SGTA, SFTPD, SFTPC, TRA2B, AKAP10, SRSF6, DUSP10, SRSF5, SRSF2, ZWINT, SLC1A2, SLC38A3, CIT, SLC27A4, SLC27A2, SLC18A1, SLC16A2, SLC11A1, SLC8A1, SLC7A2, TENT4A, SLC6A6, SLC5A2, CPSF6, SLC5A1, SLC4A1, SLC3A1, SPACA9, ADAMTS8, ADAMTS5, PRSS23, KRR1, RASSF8, PDCD10, ZHX1, SLC6A14, CCL14, CCL11, ANKRD6, CARD8, SACM1L, FAN1, CCL3, NCBP2, ATF6, SEPHS2, SCNN1A, SCAP, SCN4B, SCN3A, TRIM32, SCN2A, PAXIP1, NT5C2, SCN1B, INPP5F, DZIP1, FASTKD2, SDHA, HRH3, XPOT, NRM, USP18, TREX1, SEC14L1, ECD, SEA, SYNPO, CNKSR2, SDF2, CLCA4, IKZF3, IKZF2, ATF5, PUF60, CHSY1, CILK1, SDS, IMMT, GREM1, PAICS, STX3, TRIM3, STX1A, STK4, NPRL2, CAMKK2, YKT6, SPINT2, KHDRBS3, ST2, CD226, CGRRF1, SSR1, GNA13, DLL3, RO60, SRMS, SRI, SLC12A7, STYX, PAIP1, SLC22A1, CDC42EP3, APPBP2, HNF1A, TCEA3, DDX17, TCEA2, TCEA1, TBX5, TBP, TBL1X, TALDO1, IFI44, TAF7, SYN1, VAMP7, NPC2, GNLY, VAMP1, PRPF8, SULT1C2, OGA, NFAT5, SRD5A1, YME1L1, SNRPN, SNRPD1, CPSF4, JTB, GADD45G, EDAR, COPS8, SNAPC1, SIGLEC1, SMN2, TMED2, SMN1, SMARCC1, SMARCB1, EBNA1BP2, SLIT3, SLC22A5, COPS6, KCNQ1OT1, WDR3, C1QL1, SORD, CYSLTR1, MAP3K2, SPRR1A, PLK2, ZMYND11, ARPP21, NEK6, NCKAP1, SPOCK1, SPN, ME3, GJB6, HSPH1, SPINK1, FRS2, ALDH1L1, CD3EAP, SOX12, RUVBL2, ATXN8OS, CEP131, ATXN7, SLC7A11, RAP2A, RAP1B, PRKD3, RAG2, PPIL2, ARHGAP8, IFIT5, SHC2, RASGRP3, RAB5B, BRI3, RAB3A, TNFAIP8, TXN2, PYGM, POU2F3, PYGB, ASF1A, ARMC8, CEMIP2, RAP2B, ATXN1, EDC4, RBMY1A1, CABIN1, RBMS2, NNT, MMD, ATP6V0A2, TSPAN12, RBL1, PADI4, PRG1, CASP14, ARID4A, FAM215A, RARRES1, PHLDA3, RARS1, RUSC1, CBLC, RAPSN, PRKD2, RAD1, PAMR1, PTPRN2, PTGER3, PTGER2, PSMD7, SPAG8, PTTG3P, FBXW8, PSMD2, PLA2G2D, PSMD1, FGF21, EHF, PSMB9, PYY, PSMB8, PSMB7, PSMB5, PSMB4, PSEN1, RGS17, WSB1, TANC2, HERC4, VIRMA, TMEM158, PTPRM, PTPRK, ZDHHC5, CLIC4, PTPRG, NECTIN3, SYF2, EGFL6, RAI14, NGDN, L3MBTL1, RPAP1, PTMAP4, PTMA, TWF1, GIGYF2, PTK7, POFUT1, RENBP, ISCU, TMEM131L, RTN1, RTKN, ANKLE2, RPS27A, RPS19, RPS16, LPIN1, RFTN1, DIP2A, PHLDB1, CYFIP1, FBXL7, GSE1, ARL6IP1, MLC1, RPL36A, RPL39, RPL29, RPL24, RXRG, CUL9, RYR3, PDS5B, RAB21, SAT1, FAIM2, TSPAN31, USP33, SALL2, MYT1L, TNIK, ACSM3, KIF1B, ZHX3, VPS52, S100A12, CMTR1, HECW1, PEG10, S100A2, ARHGAP26, SIK2, PDS5A, TRAM1, RPL13, SRGAP2, RNU2-1, ECPAS, PIP5K1C, RNF6, RN7SL1, RMRP, ZFPM2, RHEB, TMED3, RYBP, GPR161, RHOQ, TARDBP, OTP, SF3B3, RHAG, RGS3, CBX6, PUM2, RNY1, ARHGEF12, MGRN1, MINAR1, OTUD3, DDHD2, CAMTA1, MCF2L, ADGRL2, ADGRL3, RPL7A, RPL5, ZNF629, NEMP1, DMXL2, DNAJC13, TUT4, NEDD4L, SYNM, ROM1, SUN1, SEMA4F, SEMA6B, TCF12, SCAF11, RASSF9, BUB3, CHAF1B, SLC16A7, GDF5, AKAP1, COIL, MSC, ST8SIA2, GCM2, GPR50, ANP32A, CACNA2D2, GPR68, DPF3, GPR55, S1PR2, CD83, CDK2AP1, ESS2, TMSB10, MACROH2A1, KDM5C, USP6, USP2, H2BC10, H2BC6, H2BC7, H2BC8, H2AC20, SLC16A6, FZD6, CNOT9, PRPF4, PRPF3, CCNB2, BRAP, ARHGEF1, ATG12, PDCD5, HGS, DYRK1B, COPS2, ALX1, TRIP4, AD5, SLBP, SEMA3B, SCLC1, KIF23, RPH3AL, ADAMTS4, ADAMTS3, ADAMTS2, LRP8, GAL3ST1, PTP4A1, MEMO1, SCAMP1, ZYX, TP53I11, ZNF236, IGDCC3, RAB3D, APBA3, PCYT1B, LAP, ITM2A, STOML1, CNOT8, RPL23, MLF2, LHX2, ITGBL1, CSRP3, RAB9A, NRXN3, LPXN, FZD3, TMPRSS11D, FADS2, CYP7B1, ST7, NCR2, MED23, MED7, ITM2B, H2BC21, DNAJA3, PKMYT1, HAT1, TMEFF1, CDKL1, PRKRA, PDLIM4, FGF17, CES2, MADD, YARS1, TCAP, GGH, CDC14A, AP3B1, PIR, BARX2, CAMK1, YBX3, AKAP4, ALDH1A2, GAS7, RGS20, NOP14, PLA2G4C, RNMT, EIF3G, EIF3D, GALNT4, B3GALT4, EIF3C, TRADD, TNFRSF25, EED, LAMTOR3, RUVBL1, ABCB11, KCNK5, AOC3, RTCA, UNC5C, TNFRSF14, PSMG1, RGS11, LDB1, MMP23A, BPY2, MMP23B, USP13, RASAL1, RPS6KA4, TNFSF18, LTBP4, SNHG3, SPARCL1, PLA2G10, CLIC3, BRSK2, PLA2G6, BAZ1B, CCRL2, TAAR5, DOK2, PIP5K1A, CLDN12, ASH2L, CLDN10, P4HA2, MGAM, KYNU, EIF2B5, PIK3R3, SGPL1, FUBP1, ZPR1, SLC5A6, GEMIN2, EIF2B4, EIF2B2, CPNE3, CUL4B, CDC42BPA, CCNA1, AP1S2, IRS4, CACNA1G, KLF11, MBD4, CUL2, ZNF224, VEZF1, TCF20, TNNC1, FAM13A, TNFAIP6, MBNL2, ABI2, TNFAIP2, WASF2, CLDN5, TMOD1, TSHZ1, ALG3, TSPAN8, MPHOSPH10, DHRS2, MPZL2, NR2E1, USPL1, SEC62, PSMD14, PRG4, CLEC3A, TNNT1, ZNF131, TOP2B, SH2D3C, DNAJB6, SMC4, UBA2, SAE1, TRAF3, TRAF1, IL18BP, DPP3, HUWE1, TPM3, GPC6, PPIF, TPH1, KIF20A, ACTR1B, RASGRP1, PDIA6, TOP3A, ANGPTL7, KLRG1, GDF11, TRIB1, IRF9, BTN3A3, CPQ, BASP1, TF, IFI30, TEP1, TEAD1, MERTK, TDGF1, ZNHIT1, TDG, TIMM44, TRBV20OR9-2, CAP2, TCN1, VAT1, STK25, ENOX2, TFE3, CACNG2, TRIM22, TJP1, TLN1, RASGRP2, TLE4, GPHN, RABEPK, SPRY2, STAM2, ZMPSTE24, STAG1, THBD, TIAL1, LILRB2, THOP1, APBB3, LANCL1, THBS4, SMYD5, TFG, TRO, TROAP, FRAT1, TM9SF4, CEP135, SAFB2, EIF5B, IPO13, VRK2, VGLL4, UBE3C, CROCC, CEP57, BEST1, USP6NL, NOS1AP, VLDLR, VIPR2, DOCK4, VCP, VCL, SLK, USH2A, ZNF516, SOCS5, MICAL2, NFE2L3, ZNF121, BABAM2, CDC42BPB, ZNF24, ZNF23, WTAP, CNBP, PITPNM1, CARTPT, WNT6, ZIC1, ZAP70, YWHAE, YES1, WNT9A, WNT7A, FEZ1, ZNF592, USF2, PIEZO1, SH2B3, UCK2, KBTBD11, PUM3, TNFSF4, HS3ST3B1, HS3ST3A1, USP3, TNFSF15, TTR, MED13, CCS, NR1D2, FGFBP1, TTF1, SLC12A6, TSTA3, EIPR1, TSPY1, TSHR, TRPC6, SEC16A, ARNT2, ATP2C2, PPP1R26, UMOD, IST1, UCHL3, ARHGEF11, TRIM14, UBE2V2, ZBTB24, HEPH, MAGI2, MRC2, TLK1, UBE2D3, TOX4, UBE2D2, RHOBTB1, NUAK1, FIG4, UBAP2L, MYEOV, PRSS8, EIF2A, LBP, LAMP2, HACE1, LAMP1, LAMB3, LAMA5, MICAL3, LAMA3, PCDH10, KIF17, KRT81, WDR48, TSHZ3, KIAA1522, KRT2, KRT1, EPB41L5, CHD8, KIF22, USP37, MAVS, NLGN4X, LRP2, LCP1, CORO1B, VARS2, SLC39A10, SELENON, FADS1, LIMK2, SENP7, ZNF304, TTYH1, RHOJ, LIG1, LHB, SPC25, SLC24A3, NDRG3, BIRC6, TBC1D24, GRAMD1B, LDHC, MIER1, KLC1, NCOA5, KIR2DL3, JUP, JAK3, ITPKA, ITIH4, C12orf10, EDA2R, ITIH2, HPSE2, IGAN1, ALX4, ITGAX, ITGAM, ITGA7, PROK2, ITGA1, CHP2, NECAB3, IRF4, TP53AIP1, SCPEP1, KCNA1, SIGIRR, CTDSP1, KIF5B, G6PC2, LSM2, CCDC181, KIF5A, MUC3B, RBPJP4, KIF3C, KCNN4, UBL5, TP53INP2, CREBZF, RPRD1B, OVOL2, OPRPN, SCAF1, KCNK1, HIVEP3, GJC2, RALGAPB, SNORD24, VPS11, SELENOS, CISD1, USE1, MEFV, MEF2A, RCC2, ADAM11, AJAP1, SLC50A1, KLHL7, MCM5, PCDHG@, PCDHB@, PCDHA@, TEX15, TEX14, RNF17, MC1R, MBD1, DCAF6, MAP3K4, LSS, DOK5, RABL6, MICB, VAC14, CCDC88A, IPO9, CD99, IQCC, ASF1B, HHAT, MGAT1, PARVA, MFAP4, INAVA, H2AJ, ZNF415, DEPDC1B, CSGALNACT1, CACNA2D3, FOXJ2, RNF20, CPXM1, MAX, PARD3, SMAD6, ENY2, SMAD5, RGMA, PARP6, NBR1, KIF15, CAPRIN1, CTNNBIP1, LYZ, CCNL1, LY75, LTBR, CYSLTR2, KMT5AP1, REXO4, CAMK1D, CD177, RTN4, DUSP22, ARNTL2, CA10, JPH1, SLC7A10, DNAJC12, MMP26, EIF5A2, MATN1, CLDND1, MATK, MAT1A, MAP2, MAGEA5, BARHL1, MAGEA12, BDH2, THAP10, MAP3K7CL, C1GALT1, NMUR2, PAK6, ERVK-6, PERP, CDH23, ALPK1, WDR26, HOXA6, CPEB4, ZKSCAN3, HNRNPF, COASY, HMGCS2, TSGA10, HMBS, ASXL3, EEPD1, DUSP16, KDM7A, HLA-E, HLA-DRB5, ILKAP, HLA-DRB3, HLA-DRA, HLA-DOB, HOXB1, HOXB2, PBLD, HKDC1, MAP9, HDAC11, GGNBP2, PRR5L, CYBRD1, SETD6, HOXD3, HOXD1, SCD5, CPED1, HOXC6, STN1, ESRP2, C10orf88, HOXC@, TRIM46, HOXB8, HOXB4, MUS81, ITFG1, SLC38A1, TMX1, GDPD5, PITPNM3, HELLS, NUF2, HARS1, HADHA, TEX101, PARP9, TRAPPC9, H2BC5, RIOK1, GZMM, MARVELD1, GUCY2D, ATAD3B, RAB34, TBC1D10A, GYPE, CDCA7, LRMDA, QTRT1, CFH, MED25, DOCK8, TDRD3, MAGED4B, TXNDC5, ACTL8, CDT1, DIAPH3, HK1, HIVEP2, PPP1R14C, CFHR1, ADAMTS12, TLR10, SLCO5A1, UBE2K, VANGL1, TRIM56, HIP1, SPRY4, CBLL1, CPSF7, CSPP1, IFNGR2, NUCKS1, ACBD3, ILF2, IL12B, DEPTOR, CYP3A43, FBXL17, NIBAN2, NABP1, PAPOLG, CENPH, PINK1, CDK15, IGHMBP2, IGHG3, IFNW1, ZFP69B, WNK2, DLK2, IMPDH1, GIGYF1, CPEB1, P3H1, MCCC2, PARVG, INPPL1, CENPK, RTP4, MAGEF1, SLC39A8, INPP5D, PDLIM2, IMPDH2, ARHGEF28, INHBB, SIL1, HHIP, CDH22, INCENP, DCLRE1C, ATG3, CUEDC2, IFNGR1, FAM124B, IFNAR1, FOXN2, NDST1, DNAJB1, RIOX1, HSPE1, MORC4, NPEPL1, TBL1XR1, HSP90AA2P, HSPA1A, RERGL, FBXO31, SHCBP1, HAND2-AS1, HRH1, ERAS, HPX, HPS1, AGBL2, HTN3, NLRX1, PPP1R3B, MMEL1, PVRIG, DDX54, IFN1@, CFI, FA2H, IDS, TMEM43, IRX3, IDH1, HTR1B, HMGN5, NKAP, ID2, ICAM4, CERS4, NAA16, IAPP, HUS1, NBPF1, RNF126, ATXN3, ABCB4, PGM3, DEF6, GEMIN4, ANO7, PGD, NEUROG3, PGAM1, TRAT1, HEBP1, PFKP, DERL2, PFKFB2, MRPS18C, PRLH, PFDN4, LAP3, RPS27L, APIP, TMX2, GALNT9, PHF2, ANAPC7, NOX3, OBP2A, ERO1A, PLA2G2A, PKLR, PKD3, SOX8, VSX1, PITX1, PIK3C3, SOCS7, EHD2, PIK3C2B, TAX1BP3, PIK3C2A, SERPINB9, RRM2B, SERPINA1, G0S2, PHKG2, PENK, CRYL1, PEG3, SH3GLB1, DNAJC27, TLR7, GMPR2, CD320, ZDHHC3, ARMCX1, SLC25A37, CDK18, PLAC8, SPG21, CDK16, DACT1, PCSK5, TAOK3, PCSK1, ZMYND10, PCNT, PRKAG2, POLK, PIPOX, NT5C3A, PHF20, LINC00328, PHF20L1, PDK4, PDHB, INSIG2, CHCHD2, PDGFB, PDE8A, SLC45A2, JPT1, CLDN18, EGFL7, DBR1, PDE4B, PDE4A, PDCD2, GPN3, NIN, ZDHHC2, SLC39A2, RHOD, NRBF2, DESI1, PKIB, GNMT, PRKACG, PRKACA, PCDH17, PRIM2, HPLH1, RND1, PRIM1, TNRC6A, PRB4, NPY4R, UBE2S, PPT1, PPP5C, PPP3R1, PCSK1N, PPP4C, STK39, PRKCE, PRKCH, INTU, MAP2K3, CHORDC1, PRSS3, CYFIP2, TPK1, PROC, ZBTB32, ACAD8, PRM1, LAMP3, CPNE7, MAPK13, MTBP, MAPK10, B3GAT1, PRKCSH, DKK4, PRKCQ, INPP5J, PPP2R5E, EML4, PLAGL2, SLCO4A1, PLXNA2, TMOD4, PLTP, CYP2S1, REPIN1, TRPM5, CNOT7, TRA2A, GPSM2, CCDC106, GPR132, DSE, PKN3, ANAPC4, DNMT3L, PLCB4, IL19, SLC25A24, PRICKLE4, PNMT, UBL3, PON2, NOB1, SLCO3A1, SLCO1B3, REM1, PPP2R1A, PPP2CA, PPP1R7, PPP1R3C, MAGEH1, NTMT1, CFAP20, PPP1CA, HIPK2, KLF15, PPID, NDUFAF4, PHPT1, PPAT, LGALSL, SNX9, SCARA3, BCAS4, NCAPG2, CDKAL1, NDUFB9, NDP, DUSP23, ADPRS, SOHLH2, SDHAF2, C1orf109, TIPIN, PIGX, NCAM2, TMEM70, MYO6, PIH1D1, CDCA4, TRMT12, SUSD4, MYO5A, MYO1D, NDUFS4, UHRF1BP1, PCBP1, ANKHD1, RRN3, MARCHF5, NFIL3, NFIC, NFIB, NFE2L1, ZRANB1, NFATC4, BTG4, NFATC1, MED18, TRIT1, DYM, GIPC2, NEFH, LRRC49, NEFM, GDPD2, ST7L, TMEM45A, ARGLU1, MYO1C, GPATCH2, MSH4, STYK1, PI4K2A, IMPACT, LGR4, LRRC59, BATF3, MNAT1, MMP12, PLXNA3, SUPT20H, TENT5A, OTUB1, MLN, PRR5, STAP2, THUMPD1, CHD7, SLC48A1, COX3, CASC1, MTM1, MAP1S, MYL4, ARHGAP17, MYH6, PNPO, MRPS18A, MMUT, MUC5AC, PLEKHG6, MUC3A, ND2, MTX1, ATAD3A, ND5, ND4, ND3, LRRC1, UBA6, OGFOD1, CRLS1, UGT1A4, PAF1, NUP88, LARP7, AZIN1, TRIM33, SIX6, EEF1AKNMT, PIGT, OPCML, CHMP3, RASD1, OGN, MPC1, OGDH, NLK, OAZ1, ACSL5, ZNF44, RAB23, RAPGEF6, GHRL, ORC4, OTX1, OTX2, REG3A, VGLL1, REV1, NAT8B, LIMA1, PARN, RTCB, TRIAP1, UFC1, ETV7, IL23A, DTL, TMEM14C, ANAPC11, PAH, P4HA1, P2RY6, OXT, RAB6B, NR4A2, RTEL1, CCNJ, ROR2, NPPB, PNP, SLC38A2, DNAJC10, SSH1, NOVA2, NOVA1, RIN2, TAF7L, CNOT3, DGCR8, NODAL, CCHCR1, EPB41L4B, UGT1A6, NKX3-1, LZTFL1, NID1, DDX56, NPR1, CYCS, NANS, BCO1, KDM3B, NTRK2, NTF4, IL17D, TM6SF1, LRP1B, NRL, STX18, PRKAG3, CHRAC1, NRDC, SLC11A2, IL20RA, FGFRL1, DUOX1, SLC37A1, SETD4, NPY5R, NRG2
-
Overgrowth Syndrome
Wikipedia
Group of rare genetic disorders involving tissue hypertrophy Overgrowth syndrome Overgrowth syndromes in children constitute a group of rare disorders that are characterised by tissue hypertrophy. ... Children with some overgrowth syndromes such as Klippel–Trénaunay–Weber syndrome can be readily detectable at birth. [3] In contrast, other overgrowth syndromes such as Proteus syndrome usually present in the postnatal period, characteristically between the second and third year of life. [2] In general, children with overgrowth syndromes are at increased risk of embryonic tumor development. Contents 1 List of overgrowth syndromes 2 See also 3 References 4 External links List of overgrowth syndromes [ edit ] Examples of overgrowth syndromes include: Beckwith–Wiedemann syndrome CLOVES syndrome Fragile X syndrome Klippel–Trénaunay–Weber syndrome Macrocephaly-capillary malformation Neurofibromatosis Proteus syndrome Simpson–Golabi–Behmel syndrome Sotos syndrome Sturge–Weber syndrome Weaver syndrome See also [ edit ] Gigantism References [ edit ] ^ Ko JM (2013). "Genetic syndromes associated with overgrowth in childhood" . ... National Cancer Institute document: "Dictionary of Cancer Terms" . v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome This oncology article is a stub .
-
Feingold Syndrome
Wikipedia
Feingold syndrome Other names Oculodigitoesophagoduodenal syndrome Feingold Syndrome is inherited in an autosomal dominant fashion. Specialty Medical genetics Feingold syndrome (also called oculodigitoesophagoduodenal syndrome ) is a rare autosomal dominant hereditary disorder . ... PMID 16906565 . ^ Celli J, van Bokhoven H, Brunner HG (November 2003). "Feingold syndrome: clinical review and genetic mapping". ... ISSN 1098-1004 . ^ "Feingold Syndrome 1 | Hereditary Ocular Diseases" . disorders.eyes.arizona.edu . ... External links [ edit ] Classification D OMIM : 164280 MeSH : C537734 C537734, C537734 DiseasesDB : 33706 External resources GeneReviews : Feingold Syndrome 1 GeneReview/NIH/UW entry on Feingold syndrome v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome v t e Genetic disorders relating to deficiencies of transcription factor or coregulators (1) Basic domains 1.2 Feingold syndrome Saethre–Chotzen syndrome 1.3 Tietz syndrome (2) Zinc finger DNA-binding domains 2.1 ( Intracellular receptor ): Thyroid hormone resistance Androgen insensitivity syndrome PAIS MAIS CAIS Kennedy's disease PHA1AD pseudohypoaldosteronism Estrogen insensitivity syndrome X-linked adrenal hypoplasia congenita MODY 1 Familial partial lipodystrophy 3 SF1 XY gonadal dysgenesis 2.2 Barakat syndrome Tricho–rhino–phalangeal syndrome 2.3 Greig cephalopolysyndactyly syndrome / Pallister–Hall syndrome Denys–Drash syndrome Duane-radial ray syndrome MODY 7 MRX 89 Townes–Brocks syndrome Acrocallosal syndrome Myotonic dystrophy 2 2.5 Autoimmune polyendocrine syndrome type 1 (3) Helix-turn-helix domains 3.1 ARX Ohtahara syndrome Lissencephaly X2 MNX1 Currarino syndrome HOXD13 SPD1 synpolydactyly PDX1 MODY 4 LMX1B Nail–patella syndrome MSX1 Tooth and nail syndrome OFC5 PITX2 Axenfeld syndrome 1 POU4F3 DFNA15 POU3F4 DFNX2 ZEB1 Posterior polymorphous corneal dystrophy Fuchs' dystrophy 3 ZEB2 Mowat–Wilson syndrome 3.2 PAX2 Papillorenal syndrome PAX3 Waardenburg syndrome 1&3 PAX4 MODY 9 PAX6 Gillespie syndrome Coloboma of optic nerve PAX8 Congenital hypothyroidism 2 PAX9 STHAG3 3.3 FOXC1 Axenfeld syndrome 3 Iridogoniodysgenesis, dominant type FOXC2 Lymphedema–distichiasis syndrome FOXE1 Bamforth–Lazarus syndrome FOXE3 Anterior segment mesenchymal dysgenesis FOXF1 ACD/MPV FOXI1 Enlarged vestibular aqueduct FOXL2 Premature ovarian failure 3 FOXP3 IPEX 3.5 IRF6 Van der Woude syndrome Popliteal pterygium syndrome (4) β-Scaffold factors with minor groove contacts 4.2 Hyperimmunoglobulin E syndrome 4.3 Holt–Oram syndrome Li–Fraumeni syndrome Ulnar–mammary syndrome 4.7 Campomelic dysplasia MODY 3 MODY 5 SF1 SRY XY gonadal dysgenesis Premature ovarian failure 7 SOX10 Waardenburg syndrome 4c Yemenite deaf-blind hypopigmentation syndrome 4.11 Cleidocranial dysostosis (0) Other transcription factors 0.6 Kabuki syndrome Ungrouped TCF4 Pitt–Hopkins syndrome ZFP57 TNDM1 TP63 Rapp–Hodgkin syndrome / Hay–Wells syndrome / Ectrodactyly–ectodermal dysplasia–cleft syndrome 3 / Limb–mammary syndrome / OFC8 Transcription coregulators Coactivator: CREBBP Rubinstein–Taybi syndrome Corepressor: HR ( Atrichia with papular lesions )
-
Catel–manzke Syndrome
Wikipedia
Unsourced material may be challenged and removed. Find sources: "Catel–Manzke syndrome" – news · newspapers · books · scholar · JSTOR ( May 2008 ) ( Learn how and when to remove this template message ) Catel–Manzke syndrome Other names Hyperphalangy-clinodactyly of index finger with Pierre Robin syndrome Catel–Manzke syndrome is a rare genetic disorder characterized by distinctive abnormalities of the index fingers; the classic features of Pierre Robin syndrome ; occasionally with additional physical findings. ... Inheritance is thought to be X-linked recessive . [1] References [ edit ] ^ Online Mendelian Inheritance in Man (OMIM): 302380 External links [ edit ] Classification D ICD - 10 : Q87.8 OMIM : 302380 MeSH : C535347 DiseasesDB : 33832 External resources Orphanet : 1388 v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome
-
Zori–stalker–williams Syndrome
Wikipedia
Genetic disease Zori–Stalker–Williams syndrome Other names Pectus excavatum, macrocephaly, short stature and dysplastic nail Zori–Stalker–Williams syndrome has an autosomal dominant pattern of inheritance . Zori–Stalker–Williams syndrome , also known as pectus excavatum, macrocephaly, short stature and dysplastic nails , [1] is a rare autosomal dominant [2] congenital disorder associated with a range of features such as pectus excavatum , macrocephaly and dysplastic nails , familial short stature, developmental delay and distinctive facies . [3] [4] Further signs are known to be associated with this syndrome. [5] The name originates from the researchers who first defined and noticed the syndrome and its clinical signs. [1] It is believed that the syndrome is inherited in an autosomal dominant pattern, though there has been no new research undertaken for this rare disease. [1] References [ edit ] ^ a b c Online Mendelian Inheritance in Man (OMIM): Pectus Excavatum, Macrocephaly, Short Stature, Dysplastic Nails - 600399 - Pectus Excavatum, Macrocephaly, Short Stature, Dysplastic Nails ^ Zori RT, Stalker HJ, Williams CA (1992). "A syndrome of familial short stature, developmental delay, pectus abnormalities, distinctive facies, and dysplastic nails". Dysmorphology and Clinical Genetics . 6 : 116–122. ^ Zori Stalker Williams syndrome at NIH 's Office of Rare Diseases ^ "Pectus excavatum macrocephaly dysplastic nails" . Orphanet . ^ ORPHANET - About rare diseases - About orphan drugs [ permanent dead link ] External links [ edit ] Classification D OMIM : 600399 MeSH : C536728 External resources Orphanet : 2835 v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome This genetic disorder article is a stub .
-
Urban–rogers–meyer Syndrome
Wikipedia
"Familial syndrome of mental retardation, short stature, contractures of the hands, and genital anomalies". ... PMID 3239569 . ^ "Urban Rogers Meyer syndrome" . Orphanet . Retrieved Aug 29, 2010 . ^ "Urban-Rogers-Meyer syndrome" . Jablonski's Syndromes Database (closed) . NLM . Retrieved Aug 29, 2010 . Further reading [ edit ] Prader – Willi habitus, osteopenia, and camptodactyly; Urban–Rogers–Meyer syndrome at NIH 's Office of Rare Diseases Jablonski's Syndromes Database: Bibliography Camera G, Marugo M, Cohen MM (Nov 1993). ... External links [ edit ] Classification D ICD - 10 : Q87.8 OMIM : 264010 MeSH : C538276 v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome This genetic disorder article is a stub .
-
Holt–oram Syndrome
Wikipedia
Holt-Oram syndrome Other names Atrio-digital syndrome, Atriodigital dysplasia Holt-Oram syndrome has an autosomal dominant pattern of inheritance Specialty Medical genetics Holt–Oram syndrome (also called atrio-digital syndrome, atriodigital dysplasia, cardiac-limb syndrome, heart-hand syndrome type 1, HOS, ventriculo-radial syndrome ) is an autosomal dominant disorder that affects bones in the arms and hands (the upper limbs) and often causes heart problems. [1] The syndrome may include an absent radial bone in the forearm, an atrial septal defect in the heart, or heart block. [2] It affects approximately 1 in 100,000 people. [2] Contents 1 Presentation 2 Genetics 3 Diagnosis 4 Treatment 5 History 6 See also 7 References 8 Further reading 9 External links Presentation [ edit ] All people with Holt-Oram syndrome have, at least one, abnormal wrist bone , which can often only be detected by X-ray . [1] Other bone abnormalities are associated with the syndrome. ... Retrieved 18 April 2018 . ^ a b c d e f g h i j k l McDermott DA, Fong JC, Basson CT. Holt-Oram Syndrome. 2004 Jul 20 [Updated 2015 Oct 8]. ... "Cardiac malformations associated with the Holt-Oram syndrome—report on a family and review of the literature". ... L.; Capobianco, G. (2016-01-01). "Holt Oram syndrome: a case report and review of the literature". ... Further reading [ edit ] GeneReview/NIH/UW entry on Holt-Oram Syndrome External links [ edit ] Classification D ICD - 10 : Q87.2 OMIM : 142900 MeSH : C535326 C535326, C535326 DiseasesDB : 5988 SNOMED CT : 205814003 External resources eMedicine : med/2940 ped/1021 GeneReviews : Holt-Oram Syndrome Orphanet : 392 v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome v t e Genetic disorders relating to deficiencies of transcription factor or coregulators (1) Basic domains 1.2 Feingold syndrome Saethre–Chotzen syndrome 1.3 Tietz syndrome (2) Zinc finger DNA-binding domains 2.1 ( Intracellular receptor ): Thyroid hormone resistance Androgen insensitivity syndrome PAIS MAIS CAIS Kennedy's disease PHA1AD pseudohypoaldosteronism Estrogen insensitivity syndrome X-linked adrenal hypoplasia congenita MODY 1 Familial partial lipodystrophy 3 SF1 XY gonadal dysgenesis 2.2 Barakat syndrome Tricho–rhino–phalangeal syndrome 2.3 Greig cephalopolysyndactyly syndrome / Pallister–Hall syndrome Denys–Drash syndrome Duane-radial ray syndrome MODY 7 MRX 89 Townes–Brocks syndrome Acrocallosal syndrome Myotonic dystrophy 2 2.5 Autoimmune polyendocrine syndrome type 1 (3) Helix-turn-helix domains 3.1 ARX Ohtahara syndrome Lissencephaly X2 MNX1 Currarino syndrome HOXD13 SPD1 synpolydactyly PDX1 MODY 4 LMX1B Nail–patella syndrome MSX1 Tooth and nail syndrome OFC5 PITX2 Axenfeld syndrome 1 POU4F3 DFNA15 POU3F4 DFNX2 ZEB1 Posterior polymorphous corneal dystrophy Fuchs' dystrophy 3 ZEB2 Mowat–Wilson syndrome 3.2 PAX2 Papillorenal syndrome PAX3 Waardenburg syndrome 1&3 PAX4 MODY 9 PAX6 Gillespie syndrome Coloboma of optic nerve PAX8 Congenital hypothyroidism 2 PAX9 STHAG3 3.3 FOXC1 Axenfeld syndrome 3 Iridogoniodysgenesis, dominant type FOXC2 Lymphedema–distichiasis syndrome FOXE1 Bamforth–Lazarus syndrome FOXE3 Anterior segment mesenchymal dysgenesis FOXF1 ACD/MPV FOXI1 Enlarged vestibular aqueduct FOXL2 Premature ovarian failure 3 FOXP3 IPEX 3.5 IRF6 Van der Woude syndrome Popliteal pterygium syndrome (4) β-Scaffold factors with minor groove contacts 4.2 Hyperimmunoglobulin E syndrome 4.3 Holt–Oram syndrome Li–Fraumeni syndrome Ulnar–mammary syndrome 4.7 Campomelic dysplasia MODY 3 MODY 5 SF1 SRY XY gonadal dysgenesis Premature ovarian failure 7 SOX10 Waardenburg syndrome 4c Yemenite deaf-blind hypopigmentation syndrome 4.11 Cleidocranial dysostosis (0) Other transcription factors 0.6 Kabuki syndrome Ungrouped TCF4 Pitt–Hopkins syndrome ZFP57 TNDM1 TP63 Rapp–Hodgkin syndrome / Hay–Wells syndrome / Ectrodactyly–ectodermal dysplasia–cleft syndrome 3 / Limb–mammary syndrome / OFC8 Transcription coregulators Coactivator: CREBBP Rubinstein–Taybi syndrome Corepressor: HR ( Atrichia with papular lesions )
-
Impossible Syndrome
Wikipedia
Unsourced material may be challenged and removed. Find sources: "Impossible syndrome" – news · newspapers · books · scholar · JSTOR ( March 2017 ) ( Learn how and when to remove this template message ) Impossible syndrome Other names Chondrodysplasia situs inversus imperforate anus polydactyly Impossible syndrome has an autosomal recessive pattern of inheritance. Impossible Syndrome , is a complex combination of human congenital malformations ( birth defects ). [1] The malformations include chondrodysplasia (improper growth of bone and cartilage), situs inversus totalis (chest and abdominal organs all a mirror image of normal), cleft larynx and epiglottis , hexadactyly (six digits) on hands and feet, diaphragmatic hernia , pancreatic abnormalities, kidney abnormal on one side and absent on the other side, micropenis and ambiguous genitalia , and imperforate anus . Only one case of Impossible Syndrome has been reported; the infant was premature and stillborn . Contents 1 Genetics 2 Diagnosis 3 References 4 External links Genetics [ edit ] The inheritance of Impossible syndrome is suspected to be autosomal recessive , which means the affected gene is located on an autosome , and two copies of the gene - one from each parent - are required to have an infant with the disorder. ... External links [ edit ] Classification D External resources Orphanet : 1424 Overview at Orphanet v t e Congenital abnormality syndromes Craniofacial Acrocephalosyndactylia Apert syndrome Carpenter syndrome Pfeiffer syndrome Saethre–Chotzen syndrome Sakati–Nyhan–Tisdale syndrome Bonnet–Dechaume–Blanc syndrome Other Baller–Gerold syndrome Cyclopia Goldenhar syndrome Möbius syndrome Short stature 1q21.1 deletion syndrome Aarskog–Scott syndrome Cockayne syndrome Cornelia de Lange syndrome Dubowitz syndrome Noonan syndrome Robinow syndrome Silver–Russell syndrome Seckel syndrome Smith–Lemli–Opitz syndrome Snyder–Robinson syndrome Turner syndrome Limbs Adducted thumb syndrome Holt–Oram syndrome Klippel–Trénaunay–Weber syndrome Nail–patella syndrome Rubinstein–Taybi syndrome Gastrulation / mesoderm : Caudal regression syndrome Ectromelia Sirenomelia VACTERL association Overgrowth syndromes Beckwith–Wiedemann syndrome Proteus syndrome Perlman syndrome Sotos syndrome Weaver syndrome Klippel–Trénaunay–Weber syndrome Benign symmetric lipomatosis Bannayan–Riley–Ruvalcaba syndrome Neurofibromatosis type I Laurence–Moon–Bardet–Biedl Bardet–Biedl syndrome Laurence–Moon syndrome Combined/other, known locus 2 ( Feingold syndrome ) 3 ( Zimmermann–Laband syndrome ) 4 / 13 ( Fraser syndrome ) 8 ( Branchio-oto-renal syndrome , CHARGE syndrome ) 12 ( Keutel syndrome , Timothy syndrome ) 15 ( Marfan syndrome ) 19 ( Donohue syndrome ) Multiple Fryns syndrome This genetic disorder article is a stub .